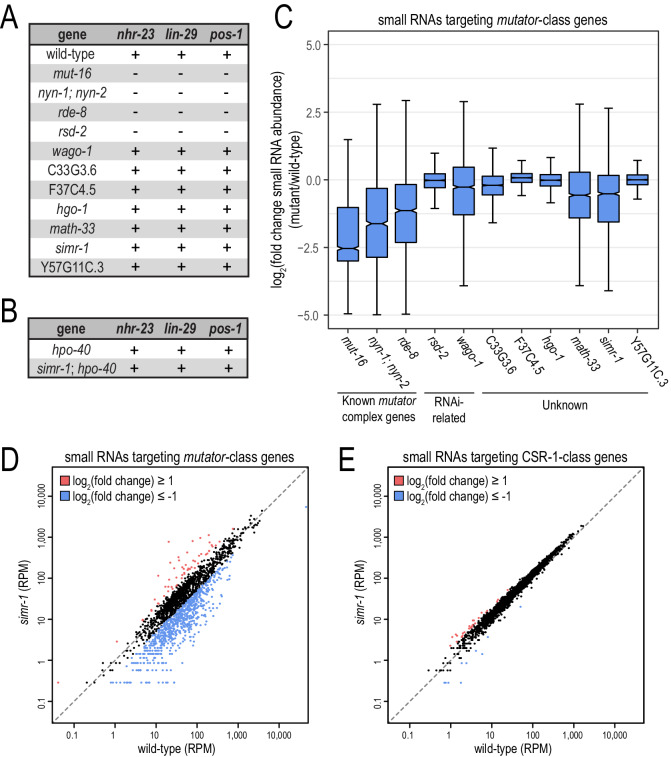
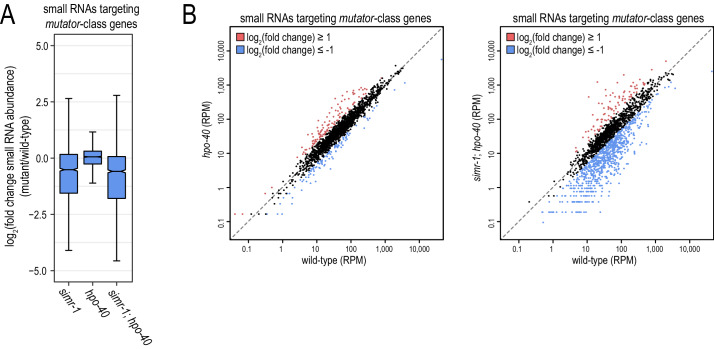
Figure 2. Small RNA-related phenotypes associated with deletions in MUT-16-associated proteins.
(A) Animals carrying deletions for each previously-uncharacterized gene identified in the MUT-16 IP-mass spec experiment were assayed for their ability to respond to somatic (nhr-23 or lin-29) or germline (pos-1) RNAi. “+” indicates wild-type response and “-” indicates RNAi-defective response. (B) Worms carrying deletions for hpo-40 single mutants or simr-1; hpo-40 double mutants were assayed for their ability to respond to somatic (nhr-23 or lin-29) or germline (pos-1) RNAi as described in (A). (C) Box plot displaying total small RNA levels targeting mutator-target genes in the indicated mutant strains relative to wild-type animals. (D,E) Scatter plots display small RNA reads per million total reads mapping to mutator-target genes (D) and CSR-1-class genes (E) in wild-type and simr-1 mutants. Genes for which log2(fold change small RNA abundance)≥1 are colored dark red and genes for which log2(fold change small RNA abundance)≤−1 are colored light blue.