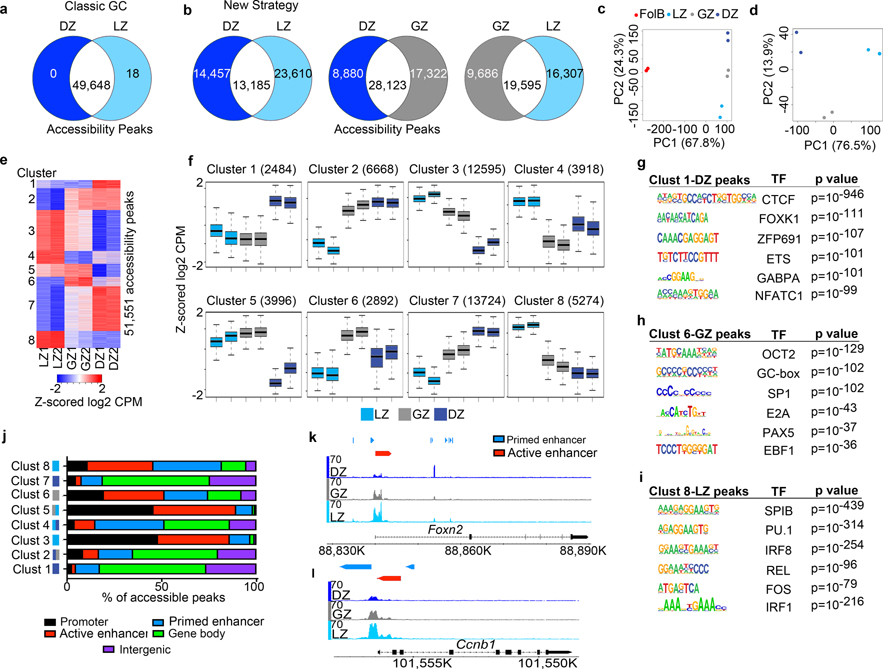
Figure 2. LZ, DZ, and GZ B cells have unique genome accessibility profiles.
a, Venn diagram comparing differentially regulated genome accessibility (ATAC-Seq) peaks between DZ and LZ B cells isolated by using Classical GC gating. q values were generated with edgeR (see methods). b, Head-to-head comparison of genome accessibility peaks among LZ, GZ, and DZ B cells isolated by the New Strategy. For (a, b), the number of common and differentially regulated accessibility peaks were determined by q<0.05 and log2 CPM>2. c and d, PCA plots displaying accessibility for indicated cell types. Each dot corresponds to an independent biological sample. e, ATAC-Seq heatmap of differentially regulated genome accessibility peaks among GCBC subsets isolated by the New Strategy. Clusters were generated by unsupervised K-means clustering. f, Box plots of differential genome accessibility at the indicated cluster of accessibility peaks. Boxes represent interquartile ranges (IQRs; Q1–Q3 percentile) and black vertical lines represent median values. Maximum and minimum values (ends of whiskers) are defined as Q3 + 1.5× the IQR and Q1 − 1.5× the IQR, respectively. g-i, TF motifs enriched in accessible regions for the indicated genome accessibility cluster and associated GCBC subset as seen for Cluster 1-DZ peaks (g), Cluster 6-GZ peaks (h), and Cluster 8-LZ peaks (i) p values were generated using Hypergeometric Optimization of Motif EnRichment (HOMER) (see methods). For each cluster, n= the number of accessibility peaks indicated in panel (f). j, Plot displaying the proportion of peaks in the indicated clusters that annotate to promoter, primed enhancer, active enhancer, gene body, and intergenic regions of the genome. Increased expression in each cluster is associated with one or more GCBC subsets, LZ-light blue, GZ-gray, and DZ-dark blue. k and l, Genome accessibility and enhancer tracks aligned at the Foxn2 (k) and Ccnb1 (l) loci for GCBCs isolated by the New Strategy. (a-l)For ATAC-Seq data, n=2 per cell type. Each n represents cells pooled from 20 mice. See also Extended Data Fig. 2 and Supplementary Data 3.