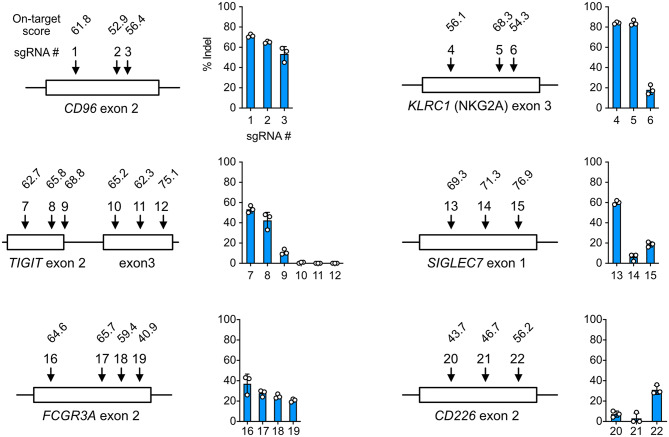
Figure 3.
Cell-based screening of efficient sgRNA. sgRNAs (22 in total) were synthesized to target the CD96, KLRC1, TIGIT, SIGLEC7, FCGR3A, and CD226 genes at the indicated exons. On-target scores of the sgRNAs were predicted in silico by the CRISPR Design tool on the Benchling website, and are indicated above the sgRNA number. A higher on-target score predicts higher editing efficiency. Percentages of insertion-or-deletion (indel) were determined by Sanger sequencing and ICE analysis. Data are shown as mean ± SD of three independent experiments. The sgRNA position and exon length are relative and not to scale.