Figure 6.
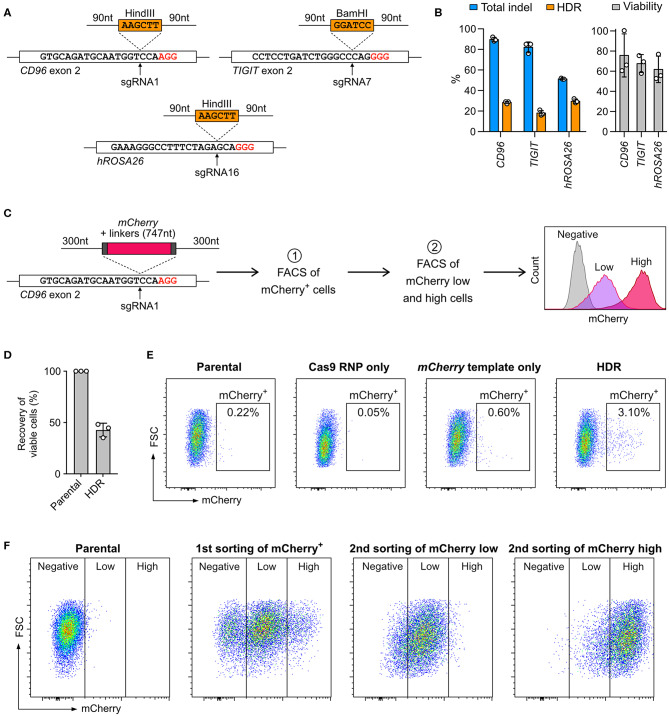
Knock-in (KI) by Cas9 RNP-mediated homology-directed repair (HDR). (A) Co-nucleofection of DNA ultramer, as HDR templates, allows the insertion of either BamHI or HindIII restriction sites at the indicated sgRNA target sites. The upstream and downstream homology arms are 90-nt long. PAM sequences are marked in red. (B) Total indels and HDR frequencies were determined by Sanger sequencing and ICE analysis. The normalized cell viability shows that single-strand DNA is less toxic than pmaxGFP to NK-92. Data are shown as mean ± SD of three independent experiments. (C) A double-stranded PCR template was used to facilitate the insertion of a promoter-less mCherry reporter gene in-frame into the CD96 exon 2 at the sgRNA1 target site. The homology arms are 300-nt long. The expression of mCherry, as driven by the endogenous CD96 promoter, was analyzed by flow cytometry. Two rounds of fluorescence-activated cell sorting (FACS) were conducted to first enrich mCherry+ cells and then further isolate mCherry low-expressing and mCherry high-expressing cells. (D) Viability assay reveals moderate level of toxicity of double-stranded PCR templates. (E) Flow cytometry plots show the percentages of mCherry+ cells at 72 h after nucleofection. (F) Distributions of mCherry low-expressing and high-expressing cells are shown after each round of cell sorting.