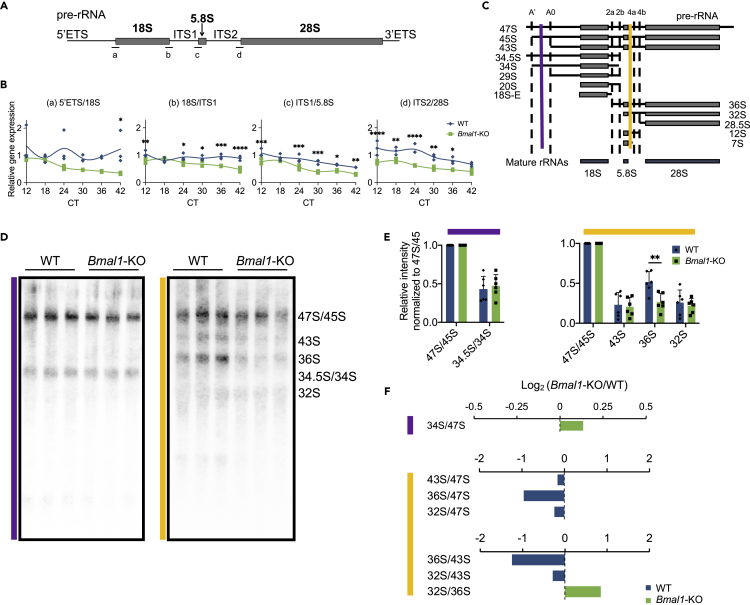
Figure 3.
Pre-ribosomal RNA Processing Is Altered in the Absence of BMAL1
(A) Schematic representation of the mammalian pre-rRNA transcript (ETS, external transcribed spacer; ITS, internal transcribed spacer). a–d indicate primer sequence positions designed for reverse transcriptase-quantitative PCR (RT-qPCR) analyses.
(B) Pre-rRNA expression profiles at indicated regions a–d from (A) in WT and Bmal1-KO MEFs harvested at circadian times (CT) 12, 18, 24, 30, 36, 42 h post-synchronization. Gene expression normalized to 18S rRNA. Individual data points are plotted. N = 3 technical replicates/time point/group, ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, and ∗∗∗∗p < 0.0001 by two-way ANOVA with Sidak's multiple comparisons test.
(C) Schematic representation of mouse pre-rRNA cleavage intermediates (laterally labeled) as depicted by Henras et al. (2015). Dotted lines delineate labeled cleavage sites (top). Vertical colored lines (purple and gold) show regions of hybridization by the Northern probes designed by Lapik et al. (2004).
(D) Northern analyses of pre-rRNA intermediates in WT and Bmal1-KO MEFs. Colored lines correspond to colored probes from (C); intermediates identified are labeled on the right. Equal amount of total RNA was loaded for analysis.
(E) Quantification of each intermediate from (D) normalized to 47S/45S. Colored lines label graphs corresponding to colored probes from (C). Data are presented as mean + SD. N = 6 biological replicates/group, ∗∗p < 0.01 by two-way ANOVA with Sidak's multiple comparisons test.
(F) Ratio Analysis of Multiple Precursors (RAMP) quantification of each intermediate from (D). Colored lines label graphs corresponding to colored probes from (C). Data are presented as the Log2 (Bmal1-KO/WT) of the mean ratio of each cleavage pair. N = 6 biological replicates/genotype.