Figure 6.
BMAL1 Links snoRNA Recruitment to Box C/D snoRNP
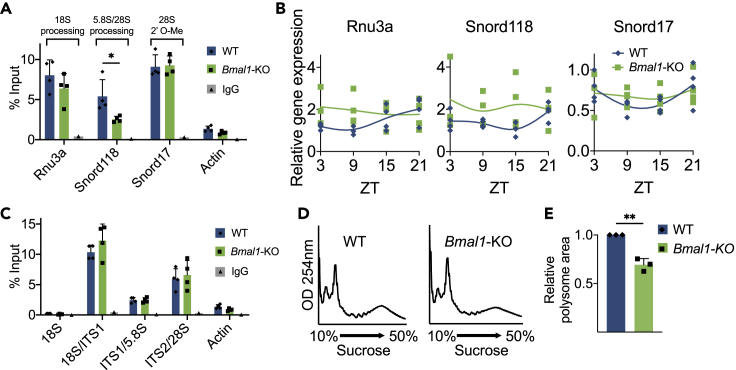
(A) RNA immunoprecipitation (RIP) using NOP58 or rabbit IgG in nucleolar fractions isolated from WT and Bmal1-KO mouse livers followed by qPCR analyses of snoRNAs: Rnu3a, Snord118, and Snord17. Actin mRNA was used as a negative control. Data are presented as mean + SD. N = 4 biological replicates/group, ∗p < 0.05 by two-way ANOVA with Tukey's multiple comparisons test.
(B) Rnu3, Snord118, and Snord17 snoRNAs expression profiles in WT and Bmal1-KO mouse livers harvested at ZT3, 9, 15, 21. Gene expression was normalized to 18S rRNA. Individual data points are plotted. N = 4 biological replicates/time point/group; no significant differences by two-way ANOVA with Sidak's multiple comparisons test.
(C) RIP using NOP58 or rabbit IgG in nucleolar fractions from WT and Bmal1-KO mouse livers followed by qPCR analysis for 18S rRNA, 18S/ITS1, ITS1/5.8S, and ITS2/28S pre-RNAs. Actin mRNA was used as a negative control. Data are presented as mean + SD. N = 4 biological replicates/group, no significant differences by two-way ANOVA with Tukey's multiple comparisons test.
(D) Polysome profiles of WT and Bmal1-KO MEFs.
(E) Quantification of the area under the polysome curve from (D). Data are presented as mean + SD. N = 3 biological replicates/group, ∗∗p < 0.01 by unpaired t test.