Abstract
Biofouling is a phenomenon that describes the fouling organisms attached to man-made surfaces immersed in water over a period of time. It has emerged as a chronic problem to the oceanic industries, especially the shipping and aquaculture fields. The metal-containing coatings that have been used for many years to prevent and destroy biofouling are damaging to the ocean and many organisms. Therefore, this calls for the critical need of natural product-based antifoulants as a substitute for its toxic counterparts. In this study, the antibacterial and antibiofilm activities of the bioactive compounds of Pseudoalteromonas sp. IBRL PD4.8 have been investigated against selected fouling bacteria. The crude extract has shown strong antibacterial activity against five fouling bacteria, with inhibition zones ranging from 9.8 to 13.7 mm and minimal inhibitory concentrations of 0.13 to 8.0 mg/ml. Meanwhile, the antibiofilm study has indicated that the extract has attenuated the initial and pre-formed biofilms of Vibrio alginolyticus FB3 by 45.37 ± 4.88% and 29.85 ± 2.56%, respectively. Moreover, micrographs from light and scanning electron microscope have revealed extensive structural damages on the treated biofilms. The active fraction was fractionated with chromatographic methods and liquid chromatography-mass spectroscopy analyses has further disclosed the presence of a polyunsaturated fatty acid 4,7,10,13-hexadecatetraenoic acid (C16H24O2). Therefore, this compound was suggested as a potential bioactive compound contributing to the antibacterial property. In conclusion, Pseudoalteromonas sp. IBRL PD4.8 is a promising source as a natural antifouling agent that can suppress the growth of five fouling bacteria and biofilms of V. alginolyticus FB3.
Key words: Pseudoalteromonas sp., antibiofilm, biofouling, liquid chromatography-mass spectroscopy, scanning electron microscope
Introduction
The aquaculture farm of the fishing industry is a severe victim from biofouling and its damaging impact. Other than the various economic consequences that had to be overcome with the cleaning, maintenance, and replacement of damaged nets, pressing focus is placed upon their detrimental effects of the cultured fishes. Colonization of bryozoans, gastropod, oyster, barnacles, and macroalgae on the pen- or cagenets have been known to implicate the fishes by causing serious water quality problems, reducing the oxygen supply, and increasing food and space competition (Fitridge et al. 2012). Furthermore, abrasion injuries, high-stress levels, and exposure to pathogenic microbes harbored by the fouling organisms have also amplified the risk of diseases to the cultured fishes (Floerl et al. 2016). Another on-going issue related to biofouling is linked to the underwater hulls of ships, where macroalgae and barnacles attach to these structures and reduce the ship’s speed due to extra drag (Townsin 2003). As a result, it increases engine stress and fuel consumption.
Nowadays, metal-containing paints like tributyltin (TBT) and copper oxide (Cu2O) have been widely utilized in the war against biofouling due to their effectiveness (Braddy 2000). However, tin and copper are released into the seawater and do not decompose rapidly, thus causing bio-accumulation in the food chain and intoxicating many lower and higher-level animals (Iwata et al. 1995; Guardiola 2012). Regardless, the most significant risk can be expected to be seen in human beings through the ingestion of contaminated seafood. Therefore, the high toxicity effect of metal-based antifoulants on non-target organisms have spurred researchers to search for the biological extracts of secondary metabolites and enzymes as a sustainable alternative (Burgess et al. 2003; Acevedo 2013). This is supplemented by the knowledge of marine organisms like corals and macroalgae that maintain their clean and foul-free surfaces by synthesizing secondary metabolites, which act as refuge mechanisms against their predators (Limna Mol et al. 2009). Nonetheless, the protection is also apparently derived by secondary metabolites produced by epiphytic bacteria that symbiotically inhabit host surfaces (Jiang et al. 2011).
According to Davey and O’Toole (2000), biofilm formation plays various roles, including environmental signaling between cells, protection from the environment, mediating in nutrient availability and metabolic activity, and attainment of gene transfer for genetic diversity. In biofouling control, biofilm, in particular, has served to be a huge hindrance as it prevents antifouling compounds from penetrating its layer and performing their inhibitory action. It has been previously discovered that a minimal inhibitory concentration (MIC) of an antimicrobial agent against biofilms increase by 1000-fold in comparison with defenseless planktonic cells (Olson et al. 2002). Therefore, the importance of getting to the root of the problem is undeniable, triggering the search for a more sustainable antifouling compound that possesses both antibacterial and antibiofilm properties.
Pseudoalteromonas sp. is a Gram-negative bacterium from the class of Gammaproteobacteria. This particular genus dominates the marine microbiome, as it is linked with bacteriolytic and algicidal properties that reveal host-protective elements (Rao et al. 2007). Hitherto, members of Pseudoalteromonas have been discovered to produce a variety of secondary metabolites boasting a broad range of bioactivities, which includes antibacterial (Isnansetyo and Kamei 2003), antifungal (Franks et al. 2006), anticancer, antimalarial, and antioxidant (Mitova et al. 2005; Martinez-Luis et al. 2011) benefits. Additionally, a brownish pigment called pyomelanin has been recently extracted from Pseudoalteromonas lipolytica, whereby its biofilms have displayed anti-larval activity against the settlement and metamorphosis of mussel Mytilus coruscus (Zeng et al. 2015; Zeng et al. 2017).
Thus, the objective of this study was to investigate the inhibitory activity of ethyl acetate extract of Pseudoalteromonas sp. IBRL PD4.8 against the fouling bacteria and biofilms of V. alginolyticus FB3. The present study, therefore, suggests the existence of bioactive compounds within the ethyl acetate extract, which are responsible for their inherent antimicrobial and antibiofilm activities.
Experimental
Materials and Methods
Bacterial strains and molecular identifications. Isolate PD4.8 was isolated from the surface of a green macroalgae, Caulerpa racemosa at Port Dickson, Malaysia. Meanwhile, the fouling bacteria (FB) were isolated from the slime layer of a fouled fish net in an aquaculture farm located in Jerejak Island, Malaysia. All strains were then grown on marine agar (MA) for 24 h at 30°C.
An analysis of 16S ribosomal DNA (rDNA) sequence were carried out to identify all strains present. The bacteria were grown in marine broth (MB) for 20 h at 150 rpm and 30°C, whereas the culture was centrifuged at 4000 rpm and a temperature of 4°C for 30 min before a pellet was extracted according to the modified phenol-chloroform extraction method (Cheng and Jiang 2006). The extracted DNA was then amplified by polymerase chain reaction (PCR) using primer 27F (5’-AGA GTT TGA TCM TGG CTC AG-3’) and reverse primer 1429R (5’-CGG TTA CCT TGT TAC GAC TT-3’). The PCR mixture was primarily consisting of 0.5 µl 27F, 0.5 µl 1492R, 0.5 µl DNA, 12.5 µl Ho Taq and 11.0 µl double distilled water (ddH2O), which was vortexed and subjected to the following PCR cycles accordingly: 94°C for 30 sec, 30 cycles of 94°C for 30 sec, 60°C for 60 seconds, 68°C for 1 min, and finally, final extension at 68°C for 5 min. Next, the DNA was semi-quantified in 0.7% (w/v) agarose gel in Tris-acetate-EDTA (TAE) buffer and visualized under ultraviolet (UV) transilluminator (BioRad) after staining with ethidium bromide. To ensure accurate sizing and approximate quantification of the DNA, a gene ruler 1 kb Plus DNA ladder was used. The PCR products were subsequently purified using the Gel Extraction Kit (Real Biotech Corporation) and sent to First Base Laboratories Sdn. Bhd. for sequencing. Afterward, the 16S rDNA sequences obtained were aligned using ClustalW of Mega Software 5.2 and then compared with sequences available in the gene bank database of National Centre for Biotechnology Information (NCBI) using Basic Local Alignment Search Tool (BLAST). A phylogenetic tree was constructed using Maximum Parsimony Method with 1000 bootstrap replications in Mega Ver.6.0 software (Tamura et al. 2013).
Fermentation and extraction. The seed culture of PD4.8 was prepared in 100 ml MB, agitated at 150 rpm and a temperature of 30°C for 24 h. Then, 10 ml of the culture with optical density (OD) 600 of 1.0 was inoculated into sterilized MB (10% v/v) and incubated at 150 rpm at a temperature of 30°C for 5 days (Bavya et al. 2011). The fermented broth was then centrifuged at 4000 rpm and a temperature of 4°C for 30 min. Next, the filtrate was extracted with ethyl acetate (EtOAc) in a separating funnel (1:1.3 v/v), whereby the resulting extract was concentrated using a rotary evaporator and kept at 4°C until further use.
Disc diffusion and minimal inhibitory concentration (MIC) assays. Twenty µl of the extract was prepared in 98% methanol (MeOH) at a concentration of 100 mg/ml and impregnated on sterilized 6 mm disc (Nor Afifah et al. 2017). Meanwhile, the FB bacteria (approx. 1 × 106 cells/ml) were prepared in 0.9% saline and swabbed evenly on the MA. The discs were then placed on the MA surface and incubated at 30°C for 24 h. Copper omadine (CuPT) (0.002 mg/disc) and 98% methanol were utilized as positive and negative controls, respectively. The resulting zones of inhibition produced around the discs were then measured in millimeter (mm).
The MIC assay of extract was undertaken according to the modified microdilution method (Yu et al. 2012). The extract, in particular, was prepared at 16.0 mg/ml (10% MeOH) and serially diluted with MB to obtain the concentrations of 8.0 mg/ml to 0.031 mg/ml. In the well, 100 µl of an extract was mixed together with 100 µl of FB, with an additional set of color control (100 µl of extract added with 100 µl MB). Two negative controls consisting of 100 µl 10% MeOH and 100 µl FB and 200 µl MB respectively were also prepared. The plate was incubated at 90 rpm at a temperature of 30°C for 24 h, whereby the well with the lowest extract concentration that showed no turbidity when compared with color control (100 µl of extract added with 100 µl MB) was taken as the MIC. All tests were done in triplicate.
Thin layer chromatography (TLC) agar-overlay assay. The crude extract was spotted at the bottom part of the TLC plate (aluminum, 2 cm × 10 cm). The plate was developed with a mixture of dichloromethane (DCM): EtOAc: MeOH (5:5:1 v/v), and subsequently observed under visible and UV lights (254 nm and 366 nm, respectively). Each spot that appeared on the TLC plate was then identified and its retention factor (Rf) calculated accordingly. Then, the TLC plate was sterilized under the UV light for 30 min and placed on the MA, with the silica surface facing upwards. Next, 10 ml of molten MA (45°C) containing 1 × 106 cells/ml of the FB was poured evenly on the TLC plate and allowed to solidify. The plate was incubated at 30°C for 24 h, and the MA surface was sprayed with 5 mg/ml ethanolic solution of p-iodonitrotetrazolium chloride (INT) post-incubation before being incubated in the dark at room temperature (28 ± 2°C) for 30 min. The bioactive spot was consequently identified via the clear zones that form against the purple background. A negative control was also used, specifically the non-spotted TLC plate.
Column chromatography and preparative TLC. To purify and collect the target bioactive fraction, a normal phase column chromatography was carried out with the isocratic elution of DCM: EtOAc: MeOH (5:5:1 v/v). The partially purified bioactive fraction was then re-tested in the MIC assay and subjected to further purification via preparative TLC using a mixture of hexane (Hex): EtOAc (1:9 v/v). The developed TLC plate was next visualized under long UV light and all sub-fractions were marked, scraped off, and soaked in 98% MeOH (HPLC) grade overnight. Then, the solutions were centrifuged at 10 000 rpm and a temperature of 4°C for 15 min, with the resulting supernatants, collected, dried, and tested in the disc diffusion assay to identify the active sub-fraction. Finally, the identified active sub-fraction was re-developed with Hex: EtOAc (1:9 v/v) so as to screen for purity.
Liquid chromatography-mass spectroscopy (LCMS). The identified bioactive sub-fraction was dissolved in 98% MeOH (HPLC grade) and filtered through a Sartorius polytetrafluoroethylene (PTFE) membrane filter (47 mm in diameter, 0.22 µm pore size). The filtered sample was then analyzed by LC coupled with the quadrupole-time-of-flight mass spectrometer (Q-TOF MS) system (Agilent Technologies). Further reverse-phase chromatography was also conducted in a Luna C18(2) column (4.6 mm × 150 mm, 5 µm, 100 Å) at a flow rate of 0.40 ml/min, with the eluent of 0.001% ammonia in deionized water (pH 7.37) and acetonitrile (6:4 v/v) over 20 min. The separated peaks were then analyzed by TOF-MS (20–2000 Da) via electrospray ionization (ESI-negative ion mode) and the consequent m/z interpreted using the MS spectra libraries (Agilent METLIN Personal Metabolite Database).
Quantitative biofilm inhibition assay. In the biofilm inhibition assay, V. alginolyticus FB3 was only tested as it produced the highest level of biofilm in comparison with other strains (data not shown). The bacterial suspension was prepared by inoculating the bacteria into 50 ml MB and incubated at 150 rpm and 30°C for 24 h. Next, the pellet was collected after centrifugation at 4000 rpm for 30 min at 4°C, and re-suspended in MB (Burmølle et al. 2006). In the initial biofilm inhibition assay, 100 µl of the bacterial suspension (OD600 = 0.15) was inoculated together with 100 µl of extract into the wells of sterilized flat bottom 96-well microtiter plate. The final concentrations of the extract are 0.03, 0.06, 0.13, 0.25, 0.5, 1.0, 2.0, 4.0, 8.0, and 16.0 mg/ml respectively. Three negative controls that comprised of 200 µl MB, 100 µl inoculum with 100 µl 5% dimethyl sulfoxide (DMSO) (v/v) in MB, and 100 µl MB with 100 µl extract, were also prepared accordingly.
The plate was incubated in a static condition for 24 h at 30°C, whereby the content of the wells after 24 hours was decanted by gently flipping the microtiter plate in a sterile container. Then, the wells were gently washed twice with the sterilized phosphate-buffer solution (PBS). The biofilms were next heat-fixed for 1 h at 60°C and stained with 210 µl of 0.06% crystal violet for 15 min. Next, the crystal violet was discarded by gently flipping the microtiter plate in a sterile container before the wells were rinsed twice with sterilized dH2O. Subsequently, the wells were flooded with 210 µl of 30% acetic acid for 10 min, following which the absorbance of the crystal violet solution was measured at 570 nm with a microtiter plate reader (Thermo Scientific).
In the pre-formed biofilm inhibition assay, the biofilm was grown by adding 100 µl of the bacterial suspension with 100 µl MB and incubated in static for 24 h. The wells were then washed with sterilized PBS before 100 µl of extract and 100 µl of fresh MB was added into them and re-incubated for 24 h at 30°C. The percentage of biofilm inhibition in comparison with the untreated control biofilms was then calculated accordingly (Nikolić et al. 2014). Meanwhile, statistical analysis was undertaken using the data obtained via Tukey’s posthoc test after one-way analysis of variance (ANOVA) was conducted using SPSS software (IBM software, version 22.0, USA). A p value of < 0.05 between the means was considered as statistically different, and all experiments were performed in four replicates accordingly:
Light microscope (LM) study. The method was undertaken as per Abu Sayem et al. (2011), with some additional modifications. Sterilized glass coverslips were placed in a tilting manner in the wells of flat-bottom 96-well microtiter plates. Then, an aliquot of 100 µl of bacterial suspension and 100 µl of extract (final concentration of 8.0 mg/ml) was added into the wells. Next, the plate was incubated in the static condition at 30°C for 24 h. For control, the extract was substituted with 5% DMSO (v/v). After the incubation process, a non-adherent biofilm was washed with PBS and the coverslip heat-fixed at 60°C for 1 h, followed with staining using 0.06% crystal violet for 15 min, and subsequent re-washing and air-drying. Finally, the stained cover slips were examined under the LM attached with a digital camera (Olympus U-CMAD3). In case of the pre-formed biofilm, the extract was introduced to the 24 h-old biofilm that grew on the coverslip.
Scanning electron microscopy (SEM) study. The SEM samples for initial and pre-formed biofilms were prepared according to the method in LM and continued further with biofilm fixing with 2.5% glutaraldehyde in PBS at 30°C for 24 h (Cai et al. 2013). The fixed biofilms were subjected to dehydration processes via incremental percentages of ethanol for 10 min each (50%, 75%, 95%, and 100% ethanol twice) before being air-dried. The coverslips were then immersed in the hexamethyldisilazane sputtered in gold and subsequently examined under the SEM (Leica Cambridge, S-360, UK).
Results
Bacteria identification. Table I shows the descriptions of all identified strains with their respective accession numbers from the NCBI database. The identified isolates were consequently denoted accordingly as Pseudoalteromonas sp. IBRL PD4.8, V. alginolyticus FB3, Pseudoalteromonas sp. FB4, Alteromonas sp. FB7, Pseudoalteromonas sp. FB9, and Bacillus sp. FB13 throughout the study. Based on the NCBI database, the epiphytic isolate Pseudoalteromonas sp. IBRL PD4.8 was closely related to Pseudoalteromonas shioyasakiensis with the 99% percentage of similarity (Table I). A result from the strict consensus tree (Fig. 1) has further supported that the epiphytic isolate is of P. shioyasakiensis with a bootstrap value of 54 %.
Table I.
Details of BLAST result, % similarity, and accession numbers of all isolates.
| Isolate | Sequence length (bp) | BLAST result | % Similarity | Accession No. |
|---|---|---|---|---|
| PD4.8 | 1385 | Pseudoalteromonas shiyosakiensis | 99 | LC131142.1 |
| FB3 | 1349 | Vibrio alginolyticus | 97 | KC884661.1 |
| FB4 | 1256 | Pseudoalteromonas sp. | 96 | JX0705058.1 |
| FB7 | 1439 | Alteromonas sp. | 93 | EF061415.1 |
| FB9 | 1398 | Pseudoalteromonas sp. | 94 | JX075059.1 |
| FB13 | 1446 | Bacillus sp. | 93 | DQ448746.1 |
Fig. 1.
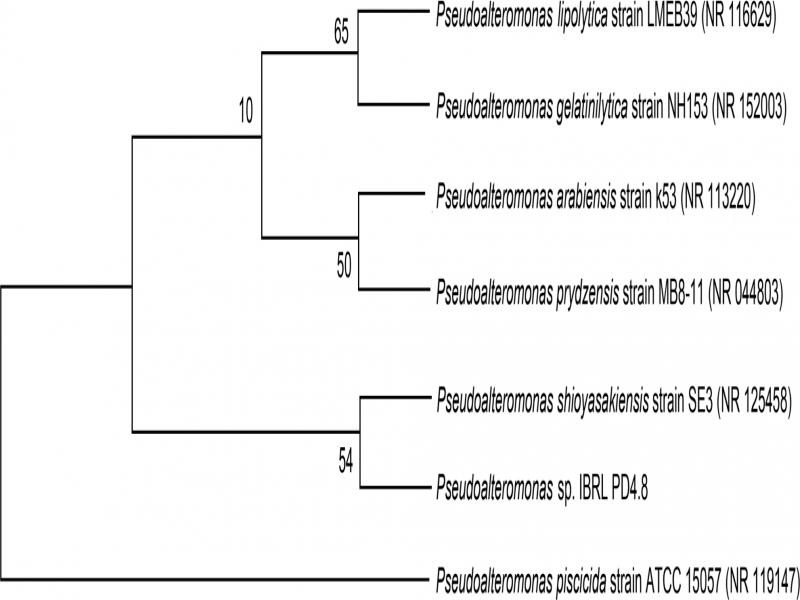
Phylogenetic tree of Pseudoalteromonas sp. IBRL PD4.8 isolated from the surface of C. racemosa.
Disc diffusion and MIC assays. At the extract concentration of 2.0 mg/disc, the ethyl acetate extract showed various degrees of inhibitory activity on the tested FB. The most susceptible FB was Bacillus sp. FB13, with an inhibition zone of 13.7 ± 2.1 mm. Meanwhile, the inhibition zones of Pseudoalteromonas sp. FB4 and Pseudoalteromonas sp. FB9 were 10.3 ± 1.5 mm and 9.0 ± 1.0 mm, respectively, whereas that of V. alginolyticus FB3 and Alteromonas sp. were 9.8 ± 1.4 mm and 11.8 ± 2.4 mm, respectively. In contrast, the extract concentration of 0.002 mg/disc was the positive control CuPT that inhibited all FB with different sizes of inhibition zones between 9.0 to 16.7 mm.
In the MIC assays, the results indicated a concentration-dependent pattern for the extract against the FB tested, whereby increased extract concentrations caused greater bacterial inhibition. This was evidenced by the clearer broth that was observed as the extract concentrations increased higher. The lowest MIC was noticed at 0.13 mg/ml against Bacillus sp. FB13. Meanwhile, for the remaining four FB (V. alginolyticus FB3, Pseudoalteromonas sp. FB4, and Alteromonas sp. FB7, Pseudoalteromonas sp. FB9) a MIC value was equal to 8.0 mg/ml (Table II).
Table II.
MIC values for crude extract and fraction F3 against fouling bacteria.
| Fouling bacteria | Crude (mg/ml), MIC | F3 (mg/ml), MIC |
|---|---|---|
| Vibrio alginolyticus FB3 | 8.00 | 8.00 |
| Pseuodoaltermonas sp. FB4 | 8.00 | 4.00 |
| Alteromonas sp. FB7 | 8.00 | 8.00 |
| Pseudoalteromonas sp. FB9 | 8.00 | 8.00 |
| Bacillus sp. FB13 | 0.13 | 0.50 |
TLC agar-overlay assay. The separation of the ethyl acetate extract on the TLC plate demonstrated one yellow spot under visible light (Rf 0.55), three spots under short UV light (Rf 0.26–0.55), and seven spots under long UV light (Rf 0.06–0.85). In contrast, TLC agar-overlay assay revealed a significant clear zone formed around the yellow spot (fluorescent green; Rf 0.55), and two fuzzy zones (light yellow; Rf 0.06 and fluorescent blue; Rf 0.35) (Fig. 2). The compounds of interest were in the yellow spot at Rf 0.55.
Fig. 2.
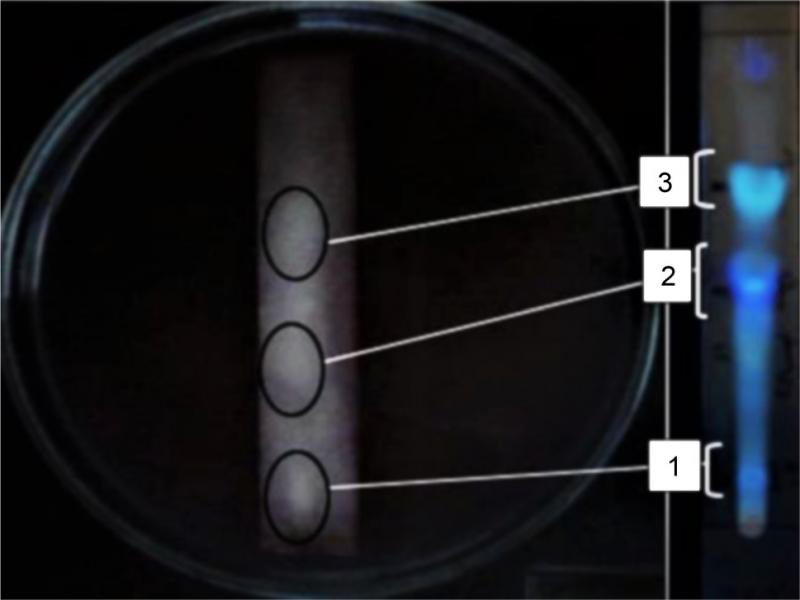
Agar-overlay assay of ethyl acetate extract of Pseudoalteromonas sp. IBRL PD4.8 against V. alginolyticus FB3.
Circle (1) light yellow, Rf 0.06; (2) fluorescent blue, Rf 0.35; (3) fluorescent green, Rf 0.55.
Antimicrobial activity and purification of fraction. Based on the agar-overlay assay (Fig. 2), three fractions were identified as potential fractions displaying antimicrobial activity. The yellow fraction (Rf 0.55) that showed the strongest inhibition zone was collected from the column chromatography. The remaining two fractions were excluded due to poor antimicrobial activity and low yield. Overall, ten fractions have been collected from the column chromatography, with the targeted yellow fraction denoted as F3. The subsequent MIC results of F3 against five FB displayed a variety of inhibitory effects, including static, decreased or enhanced activities in comparison to the crude extract (Table II). Fraction F3, in particular, has yielded a MIC value of 8.0 mg/ml against V. alginolyticus FB3, Alteromonas sp. FB7, and Pseudoalteromonas sp. FB9 similarly to the crude extract. The fraction F3 was found to be more effective towards Pseudoalteromonas sp. FB4, where the MIC was recorded to be equal to 4 mg/ml. In contrast, the MIC for fraction F3 against Bacillus sp. FB13 increased from 0.13 mg/ml to 0.50 mg/ml, indicating a reduction in antimicrobial activity.
Further separation of fraction F3 on the prepared TLC plate with the solvent system Hex: EtOAc (1:9 v/v) yielded five sub-fractions (under long UV light), namely: fluorescent green (Rf 0.55), light blue (Rf 0.71), light yellow (Rf 0.78), and light blue (Rf 0.84 and 0.90, respectively). All sub-factions on V. alginolyticus FB3 were subjected to disc diffusion assay and the sub-fraction Rf 0.55 was identified as the only sub-fraction that showed inhibitory effect against the FB tested (data not shown). It appeared as a yellow paste and was subsequently denoted as sub-fraction F3a.
LC-MS analysis. The LC-MS analysis of sub-fraction F3a identified 23 secondary metabolites within the retention times (RT) of 4.093 to 11.838 min (in the Supplementary Material). Based on the results of the searchable MS spectra libraries (Agilent METLIN Personal Metabolite Database), only six were identifiable and matched > 80% of the database search match score (score DB). The six compounds were sulfate, 4,7,10,13-hexadecatetraenoic acid, homoveratric acid, isoacitretin, sodium tetradecyl sulphate, and D1-2-Hydroxymethylethisteron. Table III summarizes the characteristics of the identified compounds. Furthermore, 12 out of the remaining 17 unidentified compounds were found to be nitrogen-containing compounds. The compound 4,7,10,13-hexadecatetraenoic acid (C16H24O2) as a type of polyunsaturated fatty acids was anticipated to be the compound responsible for the antibacterial activity of the sub-fraction F3a.
Table III.
Characteristics of compounds from LC-MS analysis of sub-fraction F3a.
| No. | RT (min) | Compound | Mass | m/z [M-H]− | Formula | Score (DB) |
|---|---|---|---|---|---|---|
| 1 | 4.233 | Sulfate | 97.967 | 96.9597 | H2O4S | 98.79 |
| 2 | 4.906 | 4,7,10,13-hexadecatetraenoic acid | 248.1763 | 293.1748 | C16H24O2 | 84.93 |
| 3 | 5.662 | Homoveratric acid | 196.0729 | 241.071 | C10H12O4 | 95.41 |
| 4 | 6.809 | Isoacitretin | 326.1891 | 325.1821 | C21H26O3 | 84.11 |
| 5 | 6.923 | Sodium tetradecyl sulfate | 294.1859 | 293.1779 | C14H30O4S | 86.66 |
| 6 | 7.617 | D1-2-Hydroxymethylethisteron | 340.2048 | 339.1978 | C22H8O3 | 85.00 |
Microtiter plate biofilm inhibition assay. Figure 3 shows the percentage of the biofilm inhibition versus the extract concentrations (0.03–16 mg/ml). At the low extract concentrations of 0.03 and 0.06 mg/ml, the initial biofilm formation was stimulated by 14.44 ± 24.73% and 4.54 ± 10.54%, respectively, which is what was demonstrated by the negative values of graph bars in Fig. 3. Furthermore, inhibition of the initial biofilm was also observed when the extract concentration increased from 0.13 mg/ml (8.17 ± 8.42%) to 0.25 mg/ml (6.22 ± 12.15%). However, it was retarded at 0.50 mg/ml before the biofilm production was re-induced (– 5.97 ± 30.14%). At 1.0 to 8.0 mg/ml, gradual increase of inhibition was observed from 23.88 ± 7.54% (1.0 mg/ml) to 45.37 ± 4.88% (8.0 mg/ml) accordingly. The biofilm inhibition was only reduced (32.32 ± 12.24%) at a concentration of 16.0 mg/ml.
Fig. 3.
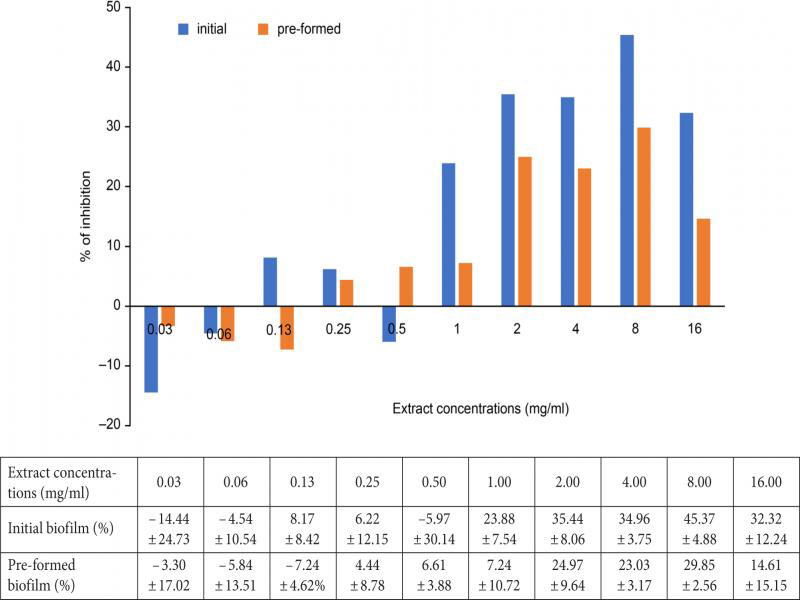
Antibiofilm activity of Pseudoalteromonas sp. IBRL PD4.8 extract against initial and pre-formed biofilm of V. alginolyticus FB3. Data of inhibition percentages are presented in the table below the figure.
Although inhibitory activity of the extract against the pre-formed biofilm was less effective in comparison with the initial biofilm, their inhibition patterns were comparable (Fig. 3). The pre-formed biofilm production was induced at lower extract concentrations of 0.03 mg/ml (– 3.30 ± 17.02%), 0.06 mg/ml (– 5.84 ± 13.51%), and 0.13 mg/ml (– 7.24 ± 4.62%). Furthermore, gradual increments in the antibiofilm activity were also observed when the extract concentrations increased from 0.25 mg/ml (4.44 ± 8.78%) to 0.50 mg/ml (6.61 ± 3.88%) and 1.0 mg/ml (7.24 ± 10.72%), accordingly. Additionally, the highest inhibition of the pre-formed biofilm was detected at 8.0 mg/ml (29.85 ± 2.56%), but it was statistically not significant (p > 0.005).
LM study. Hypothetically, the coloration of the purple image observed under LM arised from a positively charged CV stain that bounded to the envelopes of negatively charged cells and biofilm matrix. The LM results in Fig. 4 indicated that the extract was capable of inhibiting the initial and pre-formed biofilm of V. alginolyticus FB3 at 8.0 mg/ml. Meanwhile, Fig. 4a and 4c depicted the untreated sets of the initial and pre-formed biofilms, respectively. The deep purple colors from both figures were suggestive of highly dense biofilms that consisted of extracellular polymeric substances (EPS) matrix that entrapped the microcolonies. In the initial biofilm set, its exposure to the extract resulted in complete eradication of the biofilm, which left unbound cells and thin strands of damaged biofilms only (Fig. 4b). However, these results also showed that the pre-formed biofilm was less disrupted when treated with the extract at 8.0 mg/ml (Fig. 4). Some small dark patches of biofilms, in particular, have been seen to crumple together into a denser form.
Fig. 4.
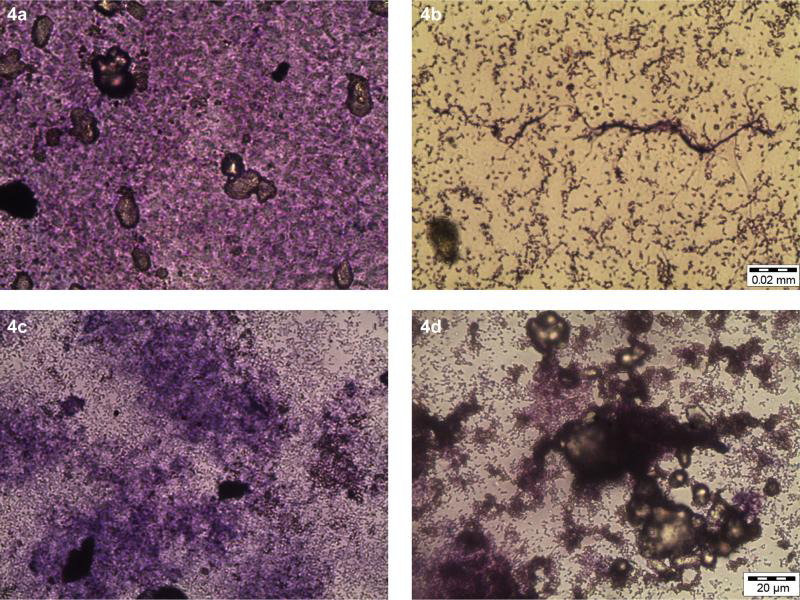
The light microscopy images of V. alginolyticus FB3 biofilms treated with Pseudoalteromonas sp. IBRL PD4.8 extract.
A. Untreated initial biofilm; B. initial biofilm treated with 8.0 mg/ml; C. untreated pre-formed biofilm; D. pre-formed biofilm treated with 8.0 mg/ml; bar = 20 µm.
SEM study. The SEM observations have revealed significant destructive effects on both initial and pre-formed biofilm architectures when treated with an extract at a concentration of 8.0 mg/ml (Fig. 5). The untreated initial biofilm shown the very low density of EPS within microcolonies that aggregated together (Fig. 5a). In contrast, the untreated pre-formed biofilm displayed more copious and non-homogenized production of EPS and microcolonies (Fig. 5c). The multilayer biofilms entrapped the microcolonies in a more complex arrangement. Moreover, the treated initial biofilm revealed that both microcolonies and EPS components alike were adversely destroyed and clumped together into unrecognized shape (Fig. 5b). Meanwhile, the treated pre-formed biofilm indicated drastic reductions with partially vanished EPS components that served to protect the cells, thereby contributing to a less compact structure (Fig. 5d).
Fig. 5.
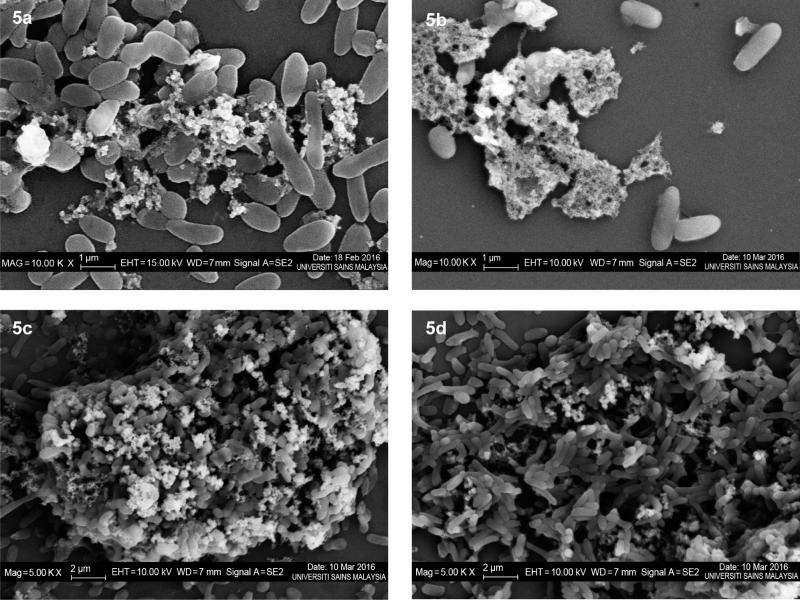
The scanning electron microscopic images of V. alginolyticus FB3 biofilms treated with the extract at a concentration of 8.0 mg/ml.
A. untreated initial biofilm (10 000×); B. treated initial biofilm (10 000×); C. untreated pre-formed biofilm (5000×); D. treated pre-formed biofilm (5000×).
Discussion
The present study was intended to investigate the inhibitory activity of the extract from Pseudoalteromonas sp. IBRL PD4.8 against fouling bacteria and biofilm production. The results have subsequently revealed that its ethyl acetate extract possesses a wide spectrum of antimicrobial activity by inhibiting both Gram-positive and Gram-negative fouling bacteria. This is the intimation that the extract may be capable of controlling complex microbial population and preventing subsequent biofouling process (Kwon et al. 2002). Only few reports have described the microbial extracts that inhibited multiple strains of fouling bacteria, and were recognized to be potential sources for the development of non-toxic antifouling compounds. One example is the ethyl acetate extract of Streptomyces filamentous R1, which was isolated from the sediment sample and inhibited three fouling bacteria (Bacillus sp. (BB11), Serratia sp. (BB13), and Alteromonas sp. (BB14)) (Bayva et al. 2011). Besides, another study has specifically depicted the fractions of ethyl acetate extract obtained from a marine fungus, Cladosporium sp. 14. It showed strong inhibitory effect and influenced the growth of three larval-settlement inducing bacteria, namely Laktonella hongkongensis, Micrococcus luteus, and Rhodovulum sp. (Qi et al. 2009).
In the disc diffusion and MIC assays, Bacillus sp. FB13 has appeared to be the most susceptible FB strain towards the extract compared to other Gram-negative FB. The less virulent property of this particular strain may be attributed to its cell wall structures, whereby it is a Gram-positive bacterium with a thick cell wall in the peptidoglycan layer and lacks the outer membrane layer. The outer membrane layer of a Gram-negative bacterium typically consists of an outer membrane with lipopolysaccharides and phospholipids, which is an effective barrier and prevents the passage of foreign molecules, including antibiotics (Nikaido 2003). Furthermore, the presence of the efflux pump within the cell envelope of the Gram-negative FB may also actively functions in expelling antibiotic molecules from its cytoplasm. This subsequently results in the reduced intracellular accumulation of antibiotics, thus minimizing the antimicrobial effect (Abdallah et al. 2007).
The antibacterial activity present in the ethyl acetate extracts of Pseudoalteromonas sp. against marine bacteria has already been discovered (Hayashida-Soiza et al. 2008; Bernbom et al. 2011). Similarly, the ethyl acetate extract of P. haloplanktis INH isolated from a scallop hatchery has also shown a powerful antibacterial activity against various marine and clinical pathogens, which includes V. alginolyticus ATCC 17749, Pseudomonas fluorescens IFO 3903, Escherichia coli IFO 3366, and Staphylococcus aureus IFO 13276. The antibacterial compound has been identified as isovaleric acid (Hayashida-Soiza et al. 2008). Moreover, three ethyl acetate extracts of Pseudoalteromonas isolates have been isolated from the deep-sea sediment of West Pacific Ocean, which also displayed antibacterial activity against several biofilmforming bacteria from Hong Kong waters (Xu et al. 2007). Similarly, two diketopiperazines identified as cyclo-(L-Ph-L-Pro) and cyclo-(L-Leu-L-Pro) have been isolated from Pseudoalteromonas sp. of octoral Leptogorgia alba (Martinez-Luis et al. 2011). At 100 µg/ml, both compounds have revealed a strong growth inhibitory effect on Vibrio sp. and B. subtillis, with the diameter of inhibitions of 14.5 to 25.0 mm.
From the LC-MS result, a polyunsaturated fatty acid named 4,7,10,13-hexadecatetraenoic acid (C16H24O2) has been predicted to be potentially responsible for the antibacterial activity against the fouling bacteria. No antimicrobial report has been found for the remaining five identified compounds (i.e. sulphate, homoveratric acid, isoacitretin, sodium tetradecyl sulphate, and D1-2-Hydroxymethylethisteron) accordingly, thus rendering them not attributed as potentially antibacterial compounds. Therefore, fatty acids have emerged as new and promising antimicrobial agents due to a strong and broad range of activities, and low likelihood of inducing bacterial resistance (Georgel et al. 2005; Desbois 2012). Previously, two short fatty acid chains called isovaleric acid (C5H10O2) and 2-methylbutyric acid (C5H10O2) have been isolated from the ethyl acetate extract of P. haloplanktis INH, and inhibited growth of six marine strains and eight clinical strains (Hayashida-Soiza et al. 2008). Besides, some polyunsaturated fatty acids with antibacterial activity have also been isolated from marine sources, such as, (6Z, 9Z, 12Z)-hexadecatrienoic acid (HTA) from a marine diatom and were effective against terrestrial and marine pathogens (Desbois 2012). Other examples also include stearidonic acid and gamma-linoleic acid sourced from the dried thalli of Enteromorpha linza, which displayed low MIC values against some oral pathogens like Candida albicans and Poryphyromonas gingivalis (Park et al. 2013).
Several antibacterial mechanisms can be proposed in elucidating the predicted 4,7,10,13-hexadecatetraenoic acid activities against susceptible fouling bacteria. The first mechanism can involve disruption of the bacterial cell membrane. The amphipathic structure and aliphatic chains of the fatty acids serve to facilitate the compound’s interaction with cell membrane components to form pores (Desbois and Smith 2010). Similarly, a toxic fatty acid like palmitoleate could create various sizes of pores at the cytoplasmic membrane of S. aureus, thus allowing the leakage of intracellular compounds (Parsons et al. 2012). Moreover, a significantly higher fatty acid concentration may cause a disastrous effect on the cell membrane via its solubilisation into fragmented parts (Desbois and Smith 2010). The second mechanism may be through the inhibition of bacterial enzymatic activity. It has been shown that lineolic acid inhibited the Fabl enzyme, which is essential for the biosynthesis of fatty acids in bacterial cell membranes (Zheng et al. 2005). Besides, the fatty acids also aid in cell lysis (Shin et al. 2007), reducing energy production in the electron transport chain system (Cartron et al. 2014), block nutrient uptake (Galbraith and Miller 1973), and peroxidation or auto-oxidation of the cells due to degraded products of the fatty acids (Wang and Johnson 1992).
In biofouling, the presence of biofilm is important to induce the settlement of larvae and spores, while also serving as a protective barrier against the antifoulant. In this study, V. alginolyticus FB3 has been selected due to the high amount of biofilm produced when compared to the other FB.
Generally, the extract obtained in this study had the ability to inhibit both the initial and pre-formed biofilms of V. alginolyticus FB3 (Fig. 3). It also shown a biphasic effect on the biofilms, since biofilm formation was induced in low extract concentration and inhibited at a higher extract concentration (Murado and Vázquez 2010). This finding is similar to that of Kaplan (2011), who demonstrated the induction of the formation of biofilm by three MRSA strains when treated with methicillin at sub-MIC concentrations. At sub-MIC, the methicillin triggered the extracellular (eDNA) and auto-aggregation mechanisms, subsequently aiding the formation processes, i.e., the initial attachment, early development, and stability retainment. Additionally, low concentrations (sub-MIC) of carbenicillin, cephaloridine, and ticaricillin have induced the expression of the cps gene, which is responsible for the synthesis of colonic acid capsular polysaccharide, and specific for mature biofilm of E. coli (Sailer et al. 2003).
Furthermore, some works have also reported the antibiofilm activities of Pseudoalteromonas species against pathogens. Strains of P. nigrifaceins and P. flavipulchra SktPp1, in particular, showed a reduction in the formation of V. cholera (Waturangi et al. 2011) and Serratia marcescens (Iqbal et al. 2015) biofilms, respectively. Meanwhile, the biofilms of S. marcescens treated with the crude extract of P. flavipulchra SktPp1 were reduced by 26.9% at a concentration of 0.1 mg/ml when compared to the control (Iqbal et al. 2015). Similarly, the filter-sterilized supernatant of Pseudoalteromonas sp. KS8 reduced the biofilm formation and the mass of mature biofilms of P. aeruginosa PAO1 by 63% and 33%, respectively (Busetti et al. 2015). The antibiofilm activity may be due to inhibition of the signaling molecule acyl-homoserine lactone (AHL) in a quorum sensing (QS) system of the P. aeruginosa PAO1. QS can be described as a process of cell-cell communication, allowing the bacteria to share information about cell density and secretion of a signaling molecule called auto-inducer signals (Miller and Bassler 2001). The system is particularly crucial in biofilm development as it permits a communication between cells to increase the population densities and consequently induce biofilm formation (Pearson et al. 1999; Lade et al. 2014). In the work by Ponnusamy et al. (2013), a significant reduction of initial biofilms of A. hydrophila has been detected under SEM and it correlated with the low amount of AHLs detected. Additionally, anti-biofilm and anti-QS activities have also been identified in the biofilm layer of S. pyogenes, which depicted the absence of dense biofilm layers upon treatment with the coral-associated bacterial extract (Thenmozhi et al. 2009).
In this study, the highest percentage of V. alginolyticus FB3 biofilm inhibition has been achieved at the concentration of the extract up to 8.0 mg/ml. Such mild inhibitory activity may be contributed to antagonistic actions of the compounds present together with the bioactive compounds. The purified compounds are generally more active than the crude extract, provided there is no synergistic enhancement within the mixture (Liu and Zhao 2016). Therefore, a bioassay-guided fractionation of the crude extract with antibiofilm activity is required to isolate the active antibiofilm compound. As it was shown in Fig. 3, the lower percentage of inhibition at a higher extract concentration (16.0 mg/ml) when compared to inhibition at 8.0 mg/ml is presumably due to the stimulation of stress-response genes of the biofilms. At that particular concentration, the extract may have become the ‘stressors’ and rendered the development of protective or adaptive stress-response, promoting biofilm formation and antimicrobial resistance (Poole 2012). One example of such stress-response is the improved number of persisters that are highly tolerant to antimicrobials (Keren et al. 2004). Persisters can be described as a small subpopulation of dormant cells that survive the treatment of bactericidal antibiotic (Lewis 2010). Nonetheless, the outcomes for every antibiofilm assay is different from one another, depending on the different types of biofilms that can be formed by the microorganisms.
Moreover, the qualitative biofilm assays such as LM and SEM provided results that indicated lower disruption of the pre-formed biofilm in comparison with the initial biofilm (Fig. 4 and 5). Based on these observations, the extract may have interfered with the initial bacterial attachment on the surface, thus preventing the formation of biofilms. This is due to bacterial attachment to a surface being the initial and fundamental step in the formation of a biofilm (O’Toole et al. 2000). Moreover, the antibacterial property of the extract may have rendered some of the planktonic cells of V. alginolyticus FB3 to be suppressed before the coverslip surface is colonized. The attachment surface may have also been modified by the extract, thus hindering bacterial colonization and biofilm development (Lewandowski and Beyenal 2014).
The glass coverslips used as the substratum is hydrophilic in nature and display high wetting capacity, in which the conditioning layer and biofilms tend to form strong adhesion (Ben Abdallah et al. 2014). In this study, the bioactive compounds in the extract may have played a role as the biosurfactant or bioemulsifier to the surface, repelling bacterial attachment and biofilm development (Neu 1996). Additionally, microbial polyphilic (also known as amphiphilic) polymers like polysaccharides and lipoteichoic acids also contain hydrophobic substituents like methyl and acyl groups, which are structurally crucial in producing a hydrophobically modified surface (Neu 1996). Therefore, the physically modified surface might have interfered with the initial bacterial adhesion and subsequently prevented biofilm formation. This amphiphilic criterion inadvertently matched the characteristic of the identified polyunsaturated fatty acid 4,7,10,13-hexadecatetrenoic acid in sub-fraction F3a, which contains a methyl group at one of its ends. Similarly, hexadecanoic acid and lineolic acid have also depicted antibiofilm activity against Vibrio spp. (Santhakumari et al. 2016), and Streptococcus mutants (Jung et al. 2014) via their surface-active compounds properties. Microscopic observations of the affected Vibrio spp. biofilms have indicated that the hexadecanoic acid reduced the initial attachment and disintegrated the mature biofilms.
In this study, SEM of the pre-formed biofilms has revealed that the biofilms were structurally thicker, hence reaffirming their function as the protective barrier that limits the entrance of antifoulants into the biofilm. Nithya and Pandian (2010) have suggested that the antibiofilm activity against the tested V. aginolyticus, V. parahaemolyticus, and V. vulnificus respectively was due to inhibition of the initial attachment, biofilm formation, and dispersion of the mature biofilm. Furthermore, LM results by Santhakumari et al. (2016) have also revealed the prevention of initial attachment and disruption of mature biofilms of V. harveyi MTCC 3438 and V. vulnificus MTCC 1145 with the extract of cynobacterial Synechococcus sp.
In the shipping industry, the removal and prevention of biofilm attachment from different ship compartments, especially the hulls and rotating discs will reduce frictional resistance. However, only a few studies have been carried out to investigate the effects of biofilm on the drag of a ship. Watanabe et al. (1969) have specifically predicted an increase of 9–10% ship resistance when slime fouling is attached to the concentric cylinders, rotating disks and a model ship. Similarly, a whopping 18% of reduced shaft horsepower is linked with propelling a ship when microbial biofilm and slight macrofouling are removed from its hull (Haslbeck and Bohlander 1992). Additionally, the composition and thickness of the biofilms can significantly influence the friction drag of a ship (Schultz and Swain 2000), as filamentous biofilms (slime and green algae) are found to cause higher skin friction. This is upon comparison with non-filamentous biofilms, thus highlighting and attributing towards biofilm complexities. All of these studies have successfully shown that biofilms can significantly increase the drag friction of a ship and reduce its speed.
In conclusion, the ethyl acetate extract of Pseudoalteromonas sp. IBRL PD4.8 contains an active compound known as 4,7,10,13-hexadecatetraenoic acid which can be a potential antibacterial compound against the fouling bacteria. It may prevent biofilm formation and eradicate established biofilm of the fouling bacteria V. alginolyticus FB3. Regardless, further investigation should be conducted to identify the specific active antibiofilm compound. Additionally, an incorporation of the extract into paint formulation should also be considered for future application purposes.
Supplementary materials are available on the journal’s website.
Acknowledgments
The authors acknowledge Universiti Sains Malaysia for the Research Grant (grant no. 1001/PBIOLOGI/815098) and Ministry of Higher Education of Malaysia for the MyBrain15 scholarship awarded.
Footnotes
Conflict of interest
The authors do not report any financial or personal connections with other persons or organizations, which might negatively affect the contents of this publication and/or claim authorship rights to this publication.
Literature
- Abu Sayem SM, Manzo E, Ciavatta L, Tramice A, Cordone A, Zanfardino A, De Felice M, Varcamonti M.. Anti-biofilm activity of an exopolysaccharide from a sponge-associated strain of Bacillus licheniformis. Microb Cell Fact. 2011;10(1):74. doi: 10.1186/1475-2859-10-74 Medline [DOI] [PMC free article] [PubMed] [Google Scholar]
- Acevedo MS, Puentes C, Carreño K, León JG, Stupak M, García M, Pérez M, Blustein G.. Antifouling paints based on marine natural products from Colombian Caribbean. Int Biodeterior Biodegradation. 2013;83:97–104. doi: 10.1016/j.ibiod.2013.05.002 [DOI] [Google Scholar]
- Bavya M, Mohanapriya P, Pazhanimurugan R, Balagurunathan R.. Potential bioactive compound from marine actinomycetes against biofouling bacteria. Indian J Geomarine Sci. 2011;40(4):578–582. [Google Scholar]
- Ben Abdallah F, Lagha R, Said K, Kallel H, Gharbi J.. Detection of cell surface hydrophobicity, biofilm and fimbirae genes in Salmonella isolated from Tunisian clinical and poultry meat. Iran J Public Health. 2014;43(4):423–431. Medline [PMC free article] [PubMed] [Google Scholar]
- Bernbom N, Ng YY, Kjelleberg S, Harder T, Gram L.. Marine bacteria from Danish coastal waters show antifouling activity against the marine fouling bacterium Pseudoalteromonas sp. strain S91 and zoospores of the green alga Ulva australis independent of bacteriocidal activity. Appl Environ Microbiol. 2011;77(24):8557–8567. doi: 10.1128/AEM.06038-11 Medline [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braddy RF., Jr. No more tin, what now for fouling control. J Protective Coating and Linings. 2000;5(6):42–46. [Google Scholar]
- Burgess JG, Boyd KG, Armstrong E, Jiang Z, Yan L, Berggren M, May U, Pisacane T, Granmo Å, Adams DR.. The development of a marine natural product-based antifouling paint. Biofouling. 2003;19(sup1) Suppl:197–205. doi: 10.1080/0892701031000061778 Medline [DOI] [PubMed] [Google Scholar]
- Burmølle M, Webb JS, Rao D, Hansen LH, Sørensen SJ, Kjelleberg S.. Enhanced biofilm formation and increased resistance to antimicrobial agents and bacterial invasion are caused by synergis tic interactions in multispecies biofilms. Appl Environ Micro biol. 2006;72(6):3916–3923. doi: 10.1128/AEM.03022-05 Medline [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busetti A, Shaw G, Megaw J, Gorman S, Maggs C, Gilmore B.. Marine-derived quorum-sensing inhibitory activities enhance the antibacterial efficacy of tobramycin against Pseudomonas aeruginosa. Mar Drugs. 2015;13(1):1–28. doi: 10.3390/md13010001 Medline [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai W, De La Fuente L, Arias CR.. Biofilm formation by the fish pathogen Flavobacterium columnare: development and parame ters affecting surface attachment. Appl Environ Microbiol. 2013;79(18):5633–5642. doi: 10.1128/AEM.01192-13 Medline [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cartron ML, England SR, Chiriac AI, Josten M, Turner R, Rauter Y, Hurd A, Sahl HG, Jones S, Foster SJ.. Bactericidal activity of the human skin fatty acid cis-6-hexadecanoic acid on Staphylococcus aureus. Antimicrob Agents Chemother. 2014;58(7):3599–3609. doi: 10.1128/AAC.01043-13 Medline [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng HR, Jiang N.. Extremely rapid extraction of DNA from bacteria and yeasts. Biotechnol Lett. 2006;28(1):55–59. doi: 10.1007/s10529-005-4688-z Medline [DOI] [PubMed] [Google Scholar]
- Davey ME, O’toole GA.. Microbial biofilms: from ecology to molecular genetics. Microbiol Mol Biol Rev. 2000;64(4):847–867. doi: 10.1128/MMBR.64.4.847-867.2000 Medline [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desbois AP, Smith VJ.. Antibacterial free fatty acids: activities, mechanisms of action and biotechnological potential. Appl Microbiol Biotechnol. 2010;85(6):1629–1642. doi: 10.1007/s00253-009-2355-3 Medline [DOI] [PubMed] [Google Scholar]
- Desbois AP. Potential applications of antimicrobial fatty acids in medicine, agriculture and other industries. Recent Pat Antiinfect Drug Discov. 2012;7(2):111–122. doi: 10.2174/157489112801619728 Medline [DOI] [PubMed] [Google Scholar]
- Fitridge I, Dempster T, Guenther J, de Nys R.. The impact and control of biofouling in marine aquaculture: a review. Biofouling. 2012;28(7):649–669. doi: 10.1080/08927014.2012.700478 Medline [DOI] [PubMed] [Google Scholar]
- Floerl O, Sunde LM, Bloecher N.. Potential environmental risks associated with biofouling management in salmon aquaculture. Aquacult Environ Interact. 2016;8:407–417. doi: 10.3354/aei00187 [DOI] [Google Scholar]
- Franks A, Egan S, Holmström C, James S, Lappin-Scott H, Kjelleberg S.. Inhibition of fungal colonization by Pseudoalteromonas tunicata provides a competitive advantage during surface colonization. Appl Environ Microbiol. 2006;72(9):6079–6087. doi: 10.1128/AEM.00559-06 Medline [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galbraith H, Miller TB.. Effect of long chain fatty acids on bacterial respiration and amino acid uptake. J Appl Bacteriol. 1973;36(4):659–675. doi: 10.1111/j.1365-2672.1973.tb04151.x Medline [DOI] [PubMed] [Google Scholar]
- Georgel P, Crozat K, Lauth X, Makrantonaki E, Seltmann H, Sovath S, Hoebe K, Du X, Rutschmann S, Jiang Z, et al. A tolllike receptor 2-responsive lipid effector pathway protects mammals against skin infections with Gram-positive bacteria. Infect Immun. 2005;73(8):4512–4521. doi: 10.1128/IAI.73.8.4512-4521.2005 Medline [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guardiola FA, Cuesta A, Meseguer J, Esteban MA.. Risks of using antifouling biocides in aquaculture. Int J Mol Sci. 2012;13(2):541–1560. doi: 10.3390/ijms13021541 Medline [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haslbeck EG, Bohlander G.. Microbial biofilm effects on drag-lab and field. In: Proceedings of the 1992 Ship Production Symposium, 2–4 September 1992 New Orleans Hyatt Regency, New Orleans, Louisiana. [Google Scholar]
- Hayashida-Soiza G, Uchida A, Mori N, Kuwahara Y, Ishida Y.. Purification and characterization of antibacterial substances produced by a marine bacterium Pseudoalteromonas haloplanktis strain. J Appl Microbiol. 2008;105(5):1672–1677. doi: 10.1111/j.1365-2672.2008.03878.x Medline [DOI] [PubMed] [Google Scholar]
- Iqbal F, Usup G, Ahmad A.. Anti-biofilm activity of Pseudoalteromonas flavipulchra SktPp1 against Serratia marcescens SMJ-11. In: Conference Proceedings of the 2015 UKM FST Postgraduate Colloquium, 15–16 April 2015 Universiti Kebangsaan Malaysia, Malaysia. [Google Scholar]
- Isnansetyo A, Kamei Y.. MC21-A, a bactericidal antibiotic produced by a new marine bacterium, Pseudoalteromonas phenolica sp. nov. O-BC30T, against methicillin-resistant Staphylococcus aureus. Anti microb Agents Chemother. 2003;47(2):480–488. doi: 10.1128/AAC.47.2.480-488.2003 Medline [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwata H, Tanabe S, Mizuno T, Tatsukawa R.. High accumulation of toxic butyltins in marine mammals from Japanese coastal waters. Environ Sci Technol. 1995;29(12):2959–2962. doi: 10.1021/es00012a011 Medline [DOI] [PubMed] [Google Scholar]
- Jiang P, Li J, Han F, Duan G, Lu X, Gu Y, Yu W.. Antibiofilm activity of an exopolysaccharide from marine bacterium Vibrio sp. QY101. PLoS One. 2011;6(4):e18514. doi: 10.1371/journal.pone.0018514 Medline [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung JE, Pandit S, Jeon JG.. Identification of linoleic acid, a main component of the n-hexane fraction from Dryopteris crassirhizoma, as an anti-Streptococcus mutans biofilm agent. Biofouling. 2014; 30(7):789–798. doi: 10.1080/08927014.2014.930446 Medline [DOI] [PubMed] [Google Scholar]
- Kaplan JB. Antibiotic-induced biofilm formation. Int J Artif Organs. 2011;34(9):737–751. doi: 10.5301/ijao.5000027 Medline [DOI] [PubMed] [Google Scholar]
- Keren I, Kaldalu N, Spoering A, Wang Y, Lewis K.. Persister cells and tolerance to antimicrobials. FEMS Microbiol Lett. 2004; 230(1):13–18. doi: 10.1016/S0378-1097(03)00856-5 Medline [DOI] [PubMed] [Google Scholar]
- Kim W, Kim Y, Kim J, Nam BH, Kim DG, An C, Lee J, Kim P, Lee H, Oh JS, et al. Liquid chromatography-mass spectrometery-based rapid secondary-metabolite profiling of marine Pseudo altero monas sp. M2. Mar Drugs. 2016;14(1):24. doi: 10.3390/md14010024 Medline [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch B, Liljefors T, Persson T, Nielsen J, Kjelleberg S, Givskov M.. The LuxR receptor: the sites of interaction with quorum-sensing signals and inhibitors. Microbiology. 2005;151(11):3589–3602. doi: 10.1099/mic.0.27954-0 Medline [DOI] [PubMed] [Google Scholar]
- Kwon KK, Lee HS, Jung S-Y, Yim J-H, Lee J-H, Lee HK.. Isolation and identification of biofilm-forming marine bacteria on glass surfaces in Dae-Ho, Korea. J Microbiol. 2002;40(4):260–266. [Google Scholar]
- Lade H, Paul D, Kweon JH.. Quorum quenching mediated approaches for control of membrane biofouling. Int J Biol Sci. 2014;10(5):550–565. doi: 10.7150/ijbs.9028 Medline [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewandowski Z, Beyenal H.. Fundamentals of biofilm research. 2014. Boca Raton (Florida): CRC Press; p. 1–61. [Google Scholar]
- Lewis K. Persister Cells. Annu Rev Microbiol. 2010;64(1):357–372. doi: 10.1146/annurev.micro.112408.134306 Medline [DOI] [PubMed] [Google Scholar]
- Limna Mol VP, Raveendran TV, Parameswaran PS.. Antifouling activity exhibited by secondary metabolites of the marine sponge, Haliclona exigua (Kirkpatrick). Int Biodeterior Biodegradation. 2009;63(1):67–72. doi: 10.1016/j.ibiod.2008.07.001 [DOI] [Google Scholar]
- Liu Y, Zhao H.. Predicting synergistic effects between compounds through their structural similarity and effects on transcriptomes. Bioinformatics. 2016;32(24):3782–3789. doi: 10.1093/bioinformatics/btw509 Medline [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez-Luis S, Ballesteros J, Gutiérrez M.. Antibacterial constituents from the octoral associated bacterium Pseudoaltero monas sp. Rev Latinoam Quím. 2011;39(1-2):75–83. [Google Scholar]
- Miller MB, Bassler BL.. Quorum sensing in bacteria. Annu Rev Microbiol. 2001;55(1):165–199. doi: 10.1146/annurev.micro.55.1.165 Medline [DOI] [PubMed] [Google Scholar]
- Mitova M, Tutino ML, Infusini G, Marino G, De Rosa S.. Exocellular peptides from Antarctic psychrophile Pseudoalteromonas haloplanktis. Mar Biotechnol (NY). 2005;7(5):523–531. doi: 10.1007/s10126-004-5098-2 Medline [DOI] [PubMed] [Google Scholar]
- Murado MA, Vázquez JA.. Biphasic toxicodynamic features of some antimicrobial agents on microbial growth: a dynamic mathema tical model and its implications on hormesis. BMC Microbiol. 2010;10:220 Medline [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neu TR. Significance of bacterial surface-active compounds in interaction of bacteria with interfaces. Microbiol Rev. 1996;60(1):151–166. Medline [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikaido H. Molecular basis of bacterial outer membrane permeability revisited. Microbiol Mol Biol Rev. 2003;67(4):593–656. doi: 10.1128/MMBR.67.4.593-656.2003 Medline [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikolić M, Vasić S, Djurdjevic J, Stefanović O, Čomić L.. Antibacterial and anti-biofilm activity of ginger (Zingiber officinale (Roscoe)) ethanolic extract. Kragujevac J Sci. 2014;36(36):129–136. doi: 10.5937/KgJSci1436129N [DOI] [Google Scholar]
- Nithya C, Pandian SK.. The in vitro antibiofilm activity of selected marine bacterial culture supernatants against Vibrio spp. Arch Microbiol. 2010;192(10):843–854. doi: 10.1007/s00203-010-0612-6 Medline [DOI] [PubMed] [Google Scholar]
- Nor Afifah S, Darah I, Sharifah Radziah MN, Wan Norhana MN, Ahmad I.. Inhibition of fouling bacteria by the marine epiphytes from selected locations in Malaysia. Mal J Sci. 2017;36(1):17–21. [Google Scholar]
- O’Toole G, Kaplan HB, Kolter R.. Biofilm formation as microbial development. Annu Rev Microbiol. 2000;54(1):49–79. doi: 10.1146/annurev.micro.54.1.49 Medline [DOI] [PubMed] [Google Scholar]
- Olson ME, Ceri H, Morck DW, Buret AG, Read RR.. Biofilm bacteria: formation and comparative susceptibility to antibiotics. Can J Vet Res. 2002;66(2):86–92. Medline [PMC free article] [PubMed] [Google Scholar]
- Park NH, Choi JS, Hwang SY, Kim YC, Hong YK, Cho K, Choi I, Choi IS.. Antimicrobial activities of stearidonic and gammalinolenic acids from the green seaweed Enteromorpha linza against several oral pathogenic bacteria. Bot Stud (Taipei, Taiwan). 2013;54(1):39. doi: 10.1186/1999-3110-54-39 Medline [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsons JB, Yao J, Frank MW, Jackson P, Rock CO.. Membrane disruption by antimicrobial fatty acids releases low-molecular-weight proteins from Staphylococcus aureus. J Bacteriol. 2012; 194(19):5294–5304. doi: 10.1128/JB.00743-12 Medline [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson JP, Van Delden C, Iglewski BH.. Active efflux and diffusion are involved in transport of Pseudomonas aeruginosa cellto-cell signals. J Bacteriol. 1999;181(4):1203–1210. Medline [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponnusamy K, Kappachery S, Thekeettle M, Song JH, Kweon JH.. Anti-biofouling property of vanillin on Aeromonas hydrophila initial biofilm on various membrane surfaces. World J Microbiol Biotechnol. 2013;29(9):1695–1703. doi: 10.1007/s11274-013-1332-2 Medline [DOI] [PubMed] [Google Scholar]
- Poole K. Stress responses as determinants of antimicrobial resis tance in Gram-negative bacteria. Trends Microbiol. 2012;20(5):227–234. doi: 10.1016/j.tim.2012.02.004 Medline [DOI] [PubMed] [Google Scholar]
- Qi SH, Xu Y, Xiong HR, Qian PY, Zhang S.. Antifouling and antibacterial compounds from a marine fungus Cladosporium sp. F14. World J Microbiol Biotechnol. 2009;25(3):399–406. doi: 10.1007/s11274-008-9904-2 [DOI] [Google Scholar]
- Rao D, Webb JS, Holmström C, Case R, Low A, Steinberg P, Kjelleberg S.. Low densities of epiphytic bacteria from the marine alga Ulva australis inhibit settlement of fouling organisms. Appl Environ Microbiol. 2007;73(24):7844–7852. doi: 10.1128/AEM.01543-07 Medline [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sailer FC, Meberg BM, Young KD.. beta-Lactam induction of colanic acid gene expression in Escherichia coli. FEMS Microbiol Lett. 2003;226(2):245–249. doi: 10.1016/S0378-1097(03)00616-5 Medline [DOI] [PubMed] [Google Scholar]
- Santhakumari S, Kannappan A, Pandian SK, Thajuddin N, Rajendran RB, Ravi AV.. Inhibitory effect of marine cyanobacterial extract on biofilm formation and virulence factor production of bacterial pathogens causing vibriosis in aquaculture. J Appl Phycol. 2016;28(1):313–324. doi: 10.1007/s10811-015-0554-0 [DOI] [Google Scholar]
- Schultz MP, Swain GW.. The influence of biofilms on skin friction drag. Biofouling. 2000;15(1-3):129–139. doi: 10.1080/08927010009386304 Medline [DOI] [PubMed] [Google Scholar]
- Shin SY, Bajpai VK, Kim HR, Kang SC.. Antibacterial activity of eicosapentaenoic acid (EPA) against foodborne and food spoilage microorganisms. Lebensm Wiss Technol. 2007;40(9):1515–1519. doi: 10.1016/j.lwt.2006.12.005 [DOI] [Google Scholar]
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S.. MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 2013;30(12):2725–2729. doi: 10.1093/molbev/mst197 Medline [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thenmozhi R, Nithyanand P, Rathna J, Karutha Pandian S.. Antibiofilm activity of coral-associated bacteria against different clinical M serotypes of Streptococcus pyogenes. FEMS Immunol Med Microbiol. 2009;57(3):284–294. doi: 10.1111/j.1574-695X.2009.00613.x Medline [DOI] [PubMed] [Google Scholar]
- Townsin RL. The ship hull fouling penalty. Biofouling. 2003; 19(sup1) Suppl:9–15. doi: 10.1080/0892701031000088535 Medline [DOI] [PubMed] [Google Scholar]
- Wang L-L, Johnson EA.. Inhibition of Listeria monocytogenes by fatty acids and monoglycerides. Appl Environ Microbiol. 1992. Feb; 58(2):624–629. Medline [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe S, Nagamatsu N, Yokoo K, Kawakami Y.. The augmentation in frictional resistance due to slime. J Kansai Soc Nav Arch. 1969;131:45–51. [Google Scholar]
- Waturangi DE, Bunardi YA, Magdalena S.. Antibiofilm activity of bacteria isolated from marine environment in Indonesia against Vibrio cholerae. Res J Microbiol. 2011;6(12):926–930. doi: 10.3923/jm.2011.926.930 [DOI] [Google Scholar]
- Xu Y, Miao L, Li XC, Xiao X, Qian PY.. Antibacterial and anti-larval activity of deep-sea bacteria from sediments of the West Pacific Ocean. Biofouling. 2007;23(2):131–137. doi: 10.1080/08927010701219323 Medline [DOI] [PubMed] [Google Scholar]
- Zeng Z, Guo XP, Cai X, Wang P, Li B, Yang JL, Wang X.. Pyomelanin from Pseudoalteromonas lipolytica reduces biofouling. Microb Biotechnol. 2017;10(6):1718–1731. doi: 10.1111/1751-7915.12773 Medline [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng Z, Guo XP, Li B, Wang P, Cai X, Tian X, Zhang S, Yang JL, Wang X.. Characterization of self-generated variants in Pseudoalteromonas lipolytica biofilm with increased antifouling activities. Appl Microbiol Biotechnol. 2015;99(23):10127–10139. doi: 10.1007/s00253-015-6865-x Medline [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng L, Chen H, Han X, Lin W, Yan X.. Antimicrobial screening and active compound isolation from marine bacterium NJ6-3-1 associated with the sponge Hymeniacidon perleve. World J Microbiol Biotechnol. 2005;21(2):201–206. doi: 10.1007/s11274-004-3318-6 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary materials are available on the journal’s website.


