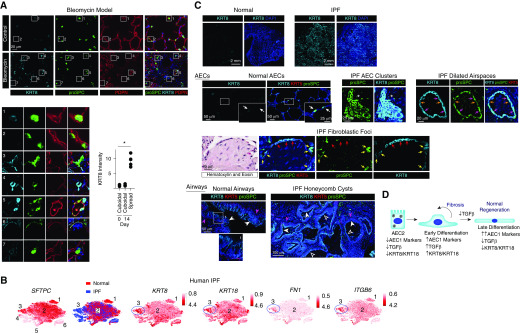
Figure 2.
Alveolar epithelial cells (AECs) in the bleomycin mouse model of pulmonary fibrosis (PF) and human idiopathic PF (IPF) have a transitional morphology and express high levels of KRT8. (A) Mice were treated with 0.025 U intratracheal bleomycin. At Day 14, lung sections were stained with the indicated antibodies. Shown are insets 1–7 enlarged. Compared with cuboidal proSPC+ PDPN− cells in the control (insets 1 and 2) and bleomycin-treated (inset 7) lungs, KRT8 is upregulated in cells with a transitional morphology and varying levels of proSPC and PDPN expression in the bleomycin-treated lungs (insets 3–6). The KRT8hi cells are found in the fibrotic areas of the bleomycin-treated lungs; KRT8 expression by type 2 AECs (AEC2s) in the uninjured areas of bleomycin-treated lungs is similar to that observed in control mice (data not shown). The fluorescence intensity of KRT8 in the cuboidal and partially spread cells was measured in control lungs and the injured areas of bleomycin-treated lungs, and the average KRT8 intensity per mouse is shown; n ≥ 4 mice per group; *P < 0.05 by Kruskal-Wallis test with Dunn’s multiple comparisons. Mean ± SEM is shown. (B) An existing dataset of single-cell RNA sequencing performed on normal human subjects and patients with PF (9) revealed the presence of six epithelial cell clusters, three of which were AEC2s. Cluster 3, which was present only in patients with IPF, displayed high expression of KRT8 and KRT18 and the TGF-β (transforming growth factor β) pathway genes FN1 (fibronectin) and ITGB6 (integrin β6). P < 10–22 for expression of these genes in cluster 3 compared with other epithelial cells. The t-distributed stochastic neighbor embedding plots were obtained from UCSC Cell Browser (https://www.nupulmonary.org/resources/?ds=fig6) on August 1, 2019. (C) Lung sections from normal human subjects (obtained from Gift of Life Michigan) or IPF explants were stained for KRT8, proSPC, and KRT5. Normal AEC2s and AEC1s expressed low levels of KRT8. White arrows indicate cuboidal AEC2s with KRT8 expression that was low or undetectable at the exposure settings used (KRT8 was detected in cuboidal AEC2s with higher exposure [data not shown]). In the IPF lung, there were clusters of cuboidal KRT8hi proSPC+ cells (asterisks). There were also dilated airspaces lined by KRT8hi KRT5− cells with a more spread morphology; most of these were proSPC+ (pink arrows), but some expressed lower levels of proSPC (orange arrows). The fibroblastic foci were covered KRT8hi KRT5− cells; most of these were proSPC− (red arrows), but some were proSPC+ (yellow arrows). Thus, these clusters, dilated airspaces, and fibroblastic foci contain hyperplastic AEC2s (some with a transitional morphology) that have upregulated KRT8. Luminal cells of the pseudostratified airways of normal lungs (solid white arrowheads) and simple (open white arrowheads) and pseudostratified (solid white arrowheads) honeycomb cysts of IPF lungs were KRT8hi, proSPC−, and KRT5−. The basal cells of normal airways and pseudostratified honeycomb cysts of IPF lungs were KRT5+ (solid pink arrowheads), consistent with previous reports (11) and were KRT8−. DAPI is represented by the dark blue stain. The images shown are representative of n ≥ 4 humans per group. Although lineage tracing of human cells cannot be performed, our data taken together with the literature suggest that the hyperplastic KRT8hi AECs that are proSPC+ and nearby proSPC− cells in the clusters, cysts, and fibroblastic foci are derived from AEC2s, whereas the KRT8hi luminal cells of the honeycomb cysts are airway derived. (D) During normal epithelial regeneration, AEC2s differentiate into AEC1s via two discrete transitional states: 1) early differentiation characterized by upregulation of AEC1 markers and KRT8 and activation of TGF-β signaling, and 2) late differentiation characterized by further upregulation of AEC1 markers, KRT8 downregulation, and deactivation of TGF-β signaling. TGF-β deactivation promotes late differentiation. In IPF, there is persistence of the TGF-β–activated, KRT8hi early differentiation state. PDPN = podoplanin.