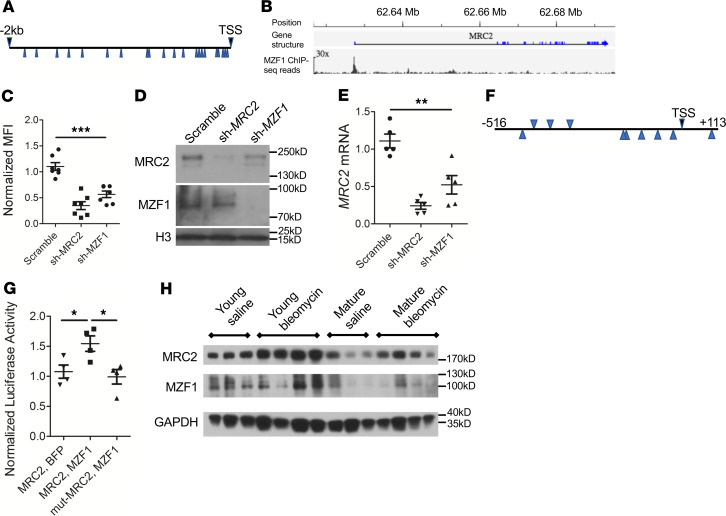
Figure 4. MZF1 regulates MRC2 expression.
(A) Schematic of 2-kb region upstream of the human MRC2 transcriptional start site with the putative MZF1 binding sites marked by blue arrows. (B) ChIP-Seq data from Encode ENCSR298QUH surrounding MRC2 gene. Scale bar: original magnification, ×30. (C) Collagen uptake assay in U937 cells treated with shRNA targeting MZF1 (sh-MZF1) or MRC2 or a scrambled control; N = 7. (D) Western blot in U937 cells treated with sh-MZF1 versus scramble or sh-MRC2 control against MRC2 and MZF1; histone H3 is a loading control (representative of N = 2). (E) Q-RT-PCR for MRC2 in sh-MZF1 cells versus scramble or sh-MRC2 control; N = 5. (F) Schematic of MZF1 putative binding sites (blue arrows) corresponding to the 625-bp peak noted in B. (G) Luciferase activity in U2OS cells treated with luciferase vector with the 625 bp from F or same sequence with 2-bp mutations in MZF1 binding sites (mut-MRC2) as well as MZF1 overexpression vector or BFP control; N = 4. (H) Representative Western blot for MRC2 and MZF1 of whole-lung lysates from young or mature mice 11 days after treatment with bleomycin or saline control; n = 3–4; a mix of male and female mice were used; GAPDH is a loading control. Statistics: (C, E, G) ANOVA. *P < 0.05, **P < 0.01, ***P < 0.001.