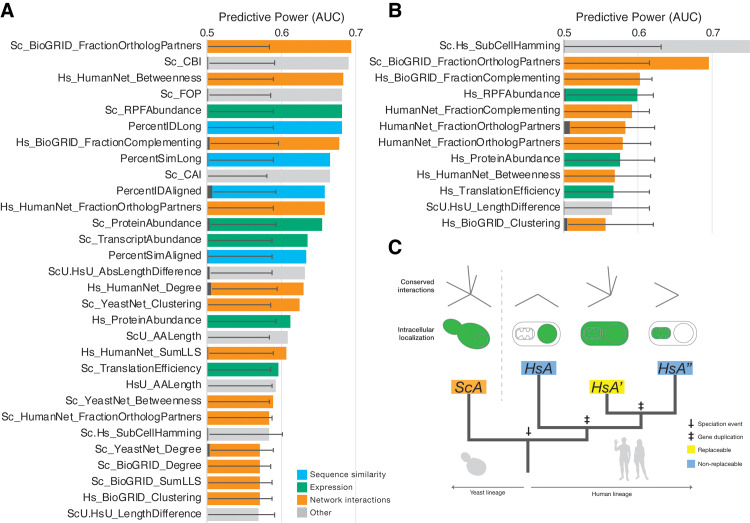
Fig 5. Replaceable 1:>2 human co-orthologs retain orthologous interaction partners and are more central in interaction networks.
(A) AUCs of the top 30 predictive features for median-collapsed informative 1:>2 co-ortholog pairs. The top two most significant features demonstrate the importance of network context in retaining ancestral functions. Specifically, human co-orthologs in highly expanded gene families that have retained a higher fraction of orthologous protein interaction partners with their yeast ortholog (FractionOrthologPartners) are more likely to replace, as well as those that maintain higher centrality in functional interaction networks (betweenness). (B) Similar to the 1:2 case, we further restricted our analysis to a subset of median-collapsed 1:>2 orthogroups that had both replaceable and nonreplaceable human co-orthologs. In this set, the subcellular localization of the human proteins appears to be predictive, in that more broadly localized co-orthologs are more likely to replace than their more organellar-specific co-orthologs. Although this AUC is not significant (indicated by being just less than 2 standard deviations off the mean), it is an obvious trend and does overtake fraction of orthologous partners as the highest performing AUC for this set. (C) Phylogenetic model of a generic 1:M orthogroup showing that the replaceable human co-ortholog has retained more orthologous interactions in a network and is localized in a similar manner to the yeast ortholog. For AUC bar plots, black overlapping bars indicate mean, and error bars indicate standard deviation for 1,000 shuffled AUC calculations for each feature. AUCs were calculated for n = 170 1:>2 ortholog pairs, collapsed to 49 and 26 median metaorthologs ([A] and [B], respectively) depending on feature availability (Materials and methods, S3 Table). The top 12 features are plotted in (B). For full source data, see S1 Data. 1:M, one-to-many; AA, amino acid; AUC, area under the receiver operating characteristic curve; CAI, codon adaptation index; CBI, codon bias index; Ens74, Ensembl genes version 74; FOP, frequency of optimal codons; Hs, H. sapiens; HsA, H. sapiens co-ortholog A; HsA′, H. sapiens co-ortholog A′; HsA′′, H. sapiens co-ortholog A′′; HsU, H. sapiens UniProt; LLS, log likelihood score; RPF, ribosome protected footprints; ScA, S. cerevisiae ortholog A; ScU, S. cerevisiae UniProt.