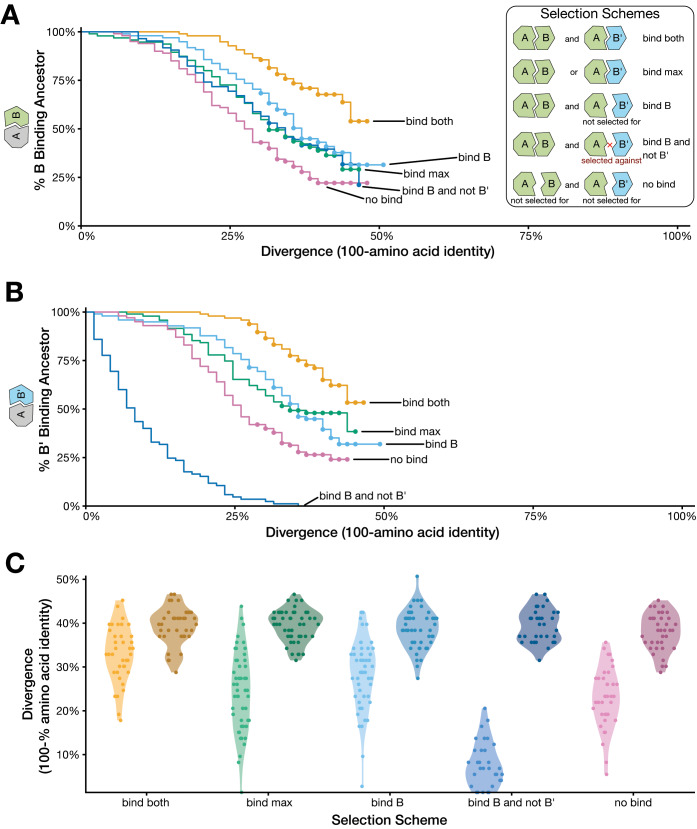
Fig 6. Simulated protein evolution suggests that diverged duplicates are less likely to bind their ancestral interaction partner.
(A) Inset: Five different types of selection scenarios were considered for a heterodimeric protein complex, considering the effects of amino acid substitutions using the Rosetta molecular modeling platform. (1) AB and AB′ (bind both). (2) AB or AB′, and only the most stable binding interface is considered (bind max). (3) AB, but AB′ was not enforced (bind B). (4) AB, and AB′ is selected against (bind B and not B′). (5) Neither AB nor AB′ is selected for (no bind). Percent of simulations in which an evolved B subunit has the ability to bind the ancestor to A. Divergence is measured by the amount B has diverged from the ancestor of B. (B) Percent of simulations in which an evolved B′ subunit has the ability to bind the ancestor to A. Divergence is measured by the amount B′ has diverged from the ancestor of B. (C) Percent divergence of duplicates when only B or B′ is able to bind the ancestor A. Lighter hues denote a duplicate that is able to bind the ancestor of A, and darker hues denote the nonbinding duplicate. Data and scripts for these figures are available at the following link: https://github.com/a-teufel/Laurent_etal_2020. Figure adapted from [35].