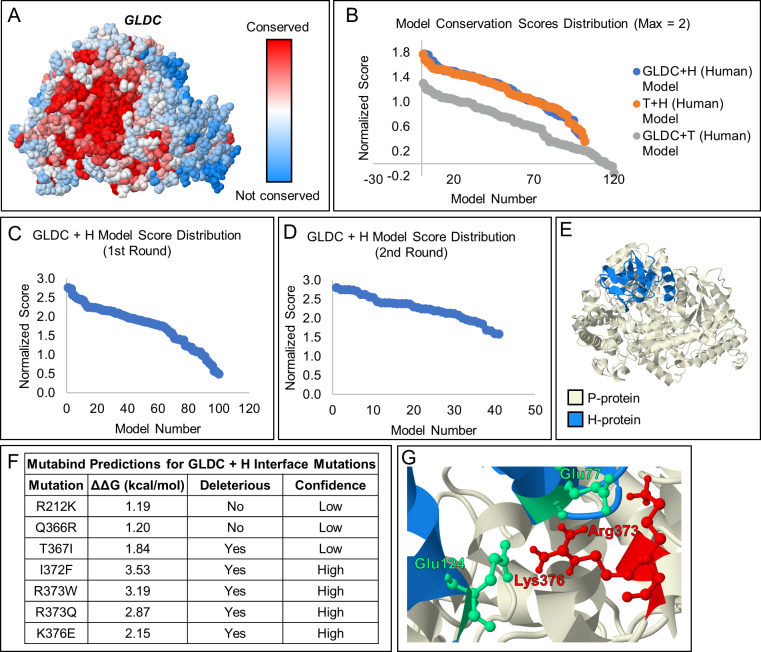
Fig 4. Model of the interaction between GLDC and H-Protein of the Glycine Cleavage System.
[A] Model showing amino acid conservation scores in GLDC calculated using the Consurf server. Highly conserved amino acids are shown in red, while poorly conserved residues are shown in blue. The highly conserved surface (deep red) found at the entry of the active site tunnel suggests this region may be a binding site for H-protein. [B] Scoring of models generated for the GLDC + H interaction (blue), T + H interaction (orange), and the GLDC + T interaction (gray). Conservation of interacting surfaces was scored from 0–1, with the most conserved surface assigned a score of 1. GLDC and T-protein likely do not interact, thus their interaction was included as a negative control. Accordingly, the GLDC + T scores were lower than the GLDC + H and H + T interaction scores, validating our novel scoring method. [C] Scoring of GLDC + H-protein interaction models scored using the conservation of interacting amino acids with the distance between the H-protein and the entry to the GLDC-protein active site tunnel as an additional parameter. H-protein was allowed to bind to any location on the GLDC-protein surface. [D] H-protein was constrained to a region of GLDC protein based on the highest scoring results of the first round of scoring, and scoring was repeated for the resulting models. [E] The proposed H- (blue) and GLDC-protein (beige) docking interaction. [F] Seven known NKH mutations were found at the predicted H-GLDC interface. The ΔΔG caused by the mutation was estimated using Mutabind. 5 of 7 mutations are predicted to caused deleterious effects to the H-GLDC protein interaction, with 3 being high confidence predictions. [G] Salt bridges between Glu77 and Glu124 of H-protein and Arg373, and Lys376 of GLDC-protein. These GLDC-protein residues are known to cause NKH when mutated.