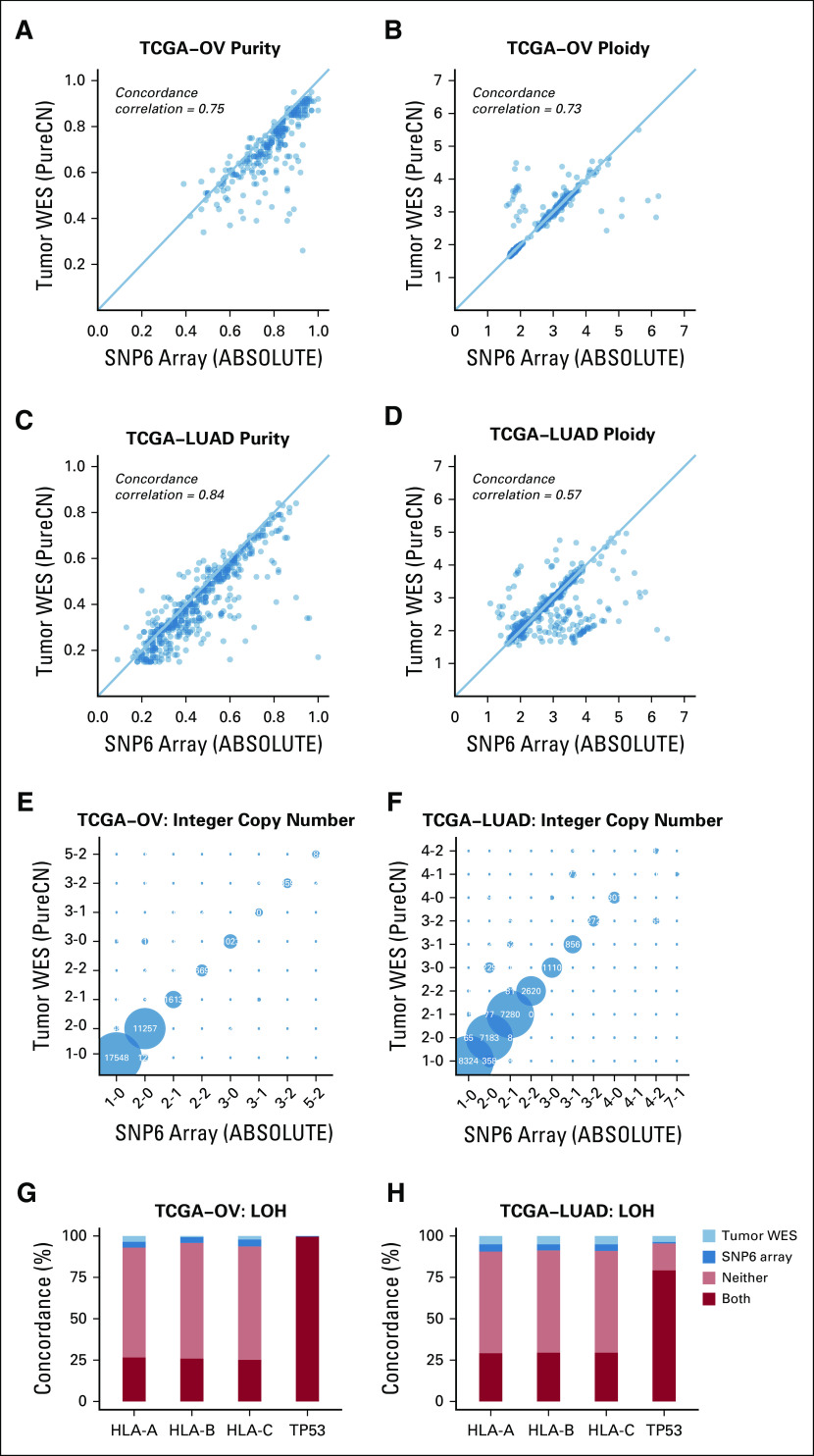
FIG 1.
Accuracy of purity, ploidy, and exome-wide copy number inference. (A-D) Comparison of purity and ploidy estimates from paired SNP6 microarray data (ABSOLUTE2) against those from tumor-only whole-exome sequencing (WES) data (PureCN) in ovarian cancer (OV) and lung adenocarcinoma (LUAD) samples. (E-F) Shown are concordances of the major and minor allele copy numbers of ABSOLUTE copy number alteration (can) calls with the corresponding tumor-only WES PureCN calls for all altered regions where both tools could make a call. Bubbles on the diagonal represent concordant calls. States where the minor copy number is 0 (1-0, 2-0, 3-0, 4-0) are regions in loss of heterozygosity (LOH). (G-H) Concordance of LOH calls between the 2 analyses was further reviewed on HLA-A/B/C and TP53 loci for the cases with sufficient power to detect LOH: LOH observed in both microarray and WES analyses (both, dark red); absent in both analyses (neither, orange); detected only from microarray data (SNP6 array, dark blue); and detected only from WES data (tumor WES, light blue). TCGA, The Cancer Genome Atlas.