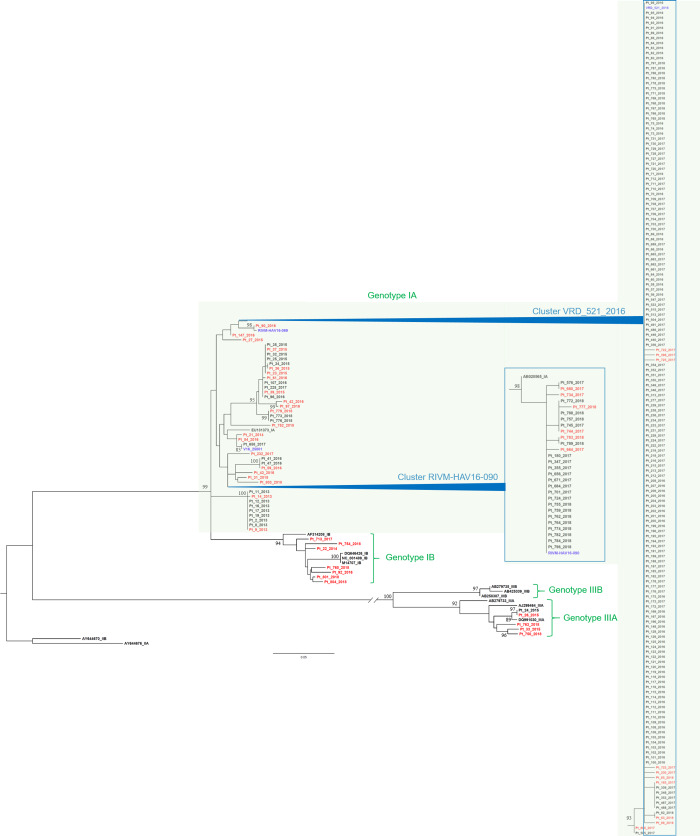
Fig 1. Maximum-likelihood phylogenetic analysis of all the sequences obtained until October 2018 from Lazio region.
Phylogenetic Maximum-Likelihood tree, built with a total of 246 459 nt-long sequences encompassing the VP1X2A junction region of HAV genome, based on the maximum-likelihood method with the General Time Reversible model + G. All the sequences obtained until October 2018 from Lazio region are included. Moreover, the tree includes 16 reference sequences from GenBank (genotype IA: EU131373; AB020565.1; genotype IB: M14707; DQ646426; NC001489; AF314208; genotype IIA: AY644676; genotype IIB: AY644670; genotype IIIA: AJ299464; DQ991030; AB279733; genotype IIIB: AB279735; AB425339; AB258387), and four sequences (VRD_521_2016 and RIVM-HAV16-90, RIVM-HAV16-69 and V16_25801, in blue) associated with epidemic clusters among MSM. The bar represents the genetic distance (substitution per nucleotide position). Bootstrap analysis with 10,000 replicates was performed to assess the significance of the nodes; values greater than 80.