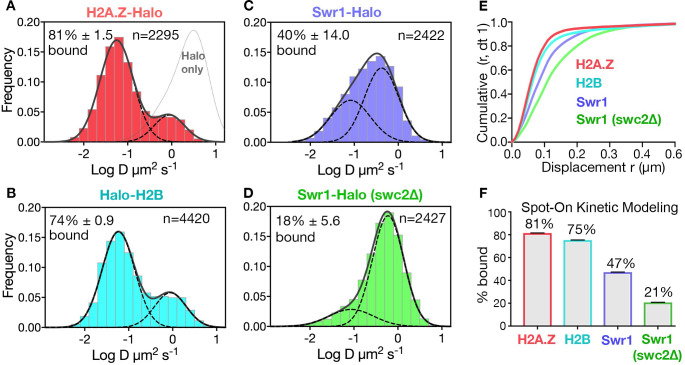
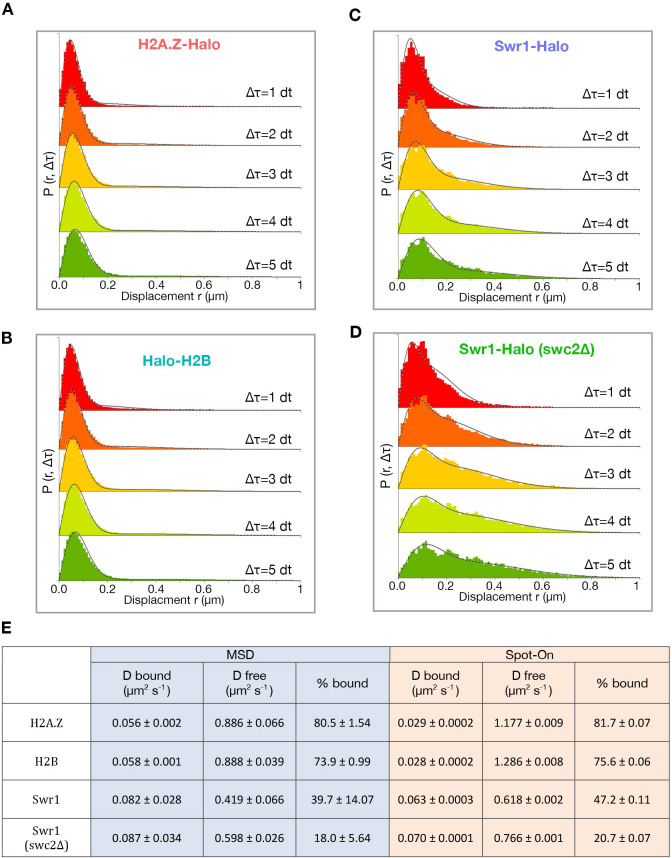
Figure 1. Diffusive behaviors of protein fusions to HaloTag (Halo) reveal chromatin-bound and free populations in live yeast.
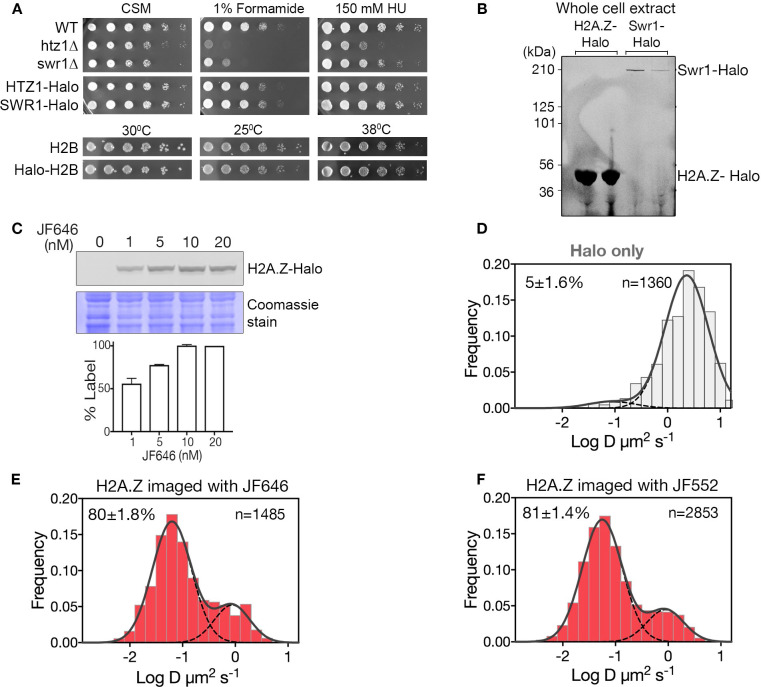
(A, B) Normalized histograms and two-component Gaussian fits for H2A.Z-Halo (A) and Halo-H2B (B) show the log diffusion coefficient distributions. The Gaussian fit for HaloTag is shown for reference (‘Halo only’ in A). (C, D) Normalized histograms and two-component Gaussian fits for Swr1-Halo in WT cells (C) and the swc2Δ mutant (D). Solid line: sum of two-component fit; dashed line: individual component. Percent value of the slow component along with Bootstrap resampling errors and the number of trajectories (n) are indicated. (E) Cumulative distribution functions (CDF) of 10 ms displacements. (F) Spot-On results with fitting errors showing fractions of chromatin-bound molecules derived from modeling CDFs over 10–50 ms intervals. All molecules tracked with JF646 dye except Halo only, which was imaged with JF552.