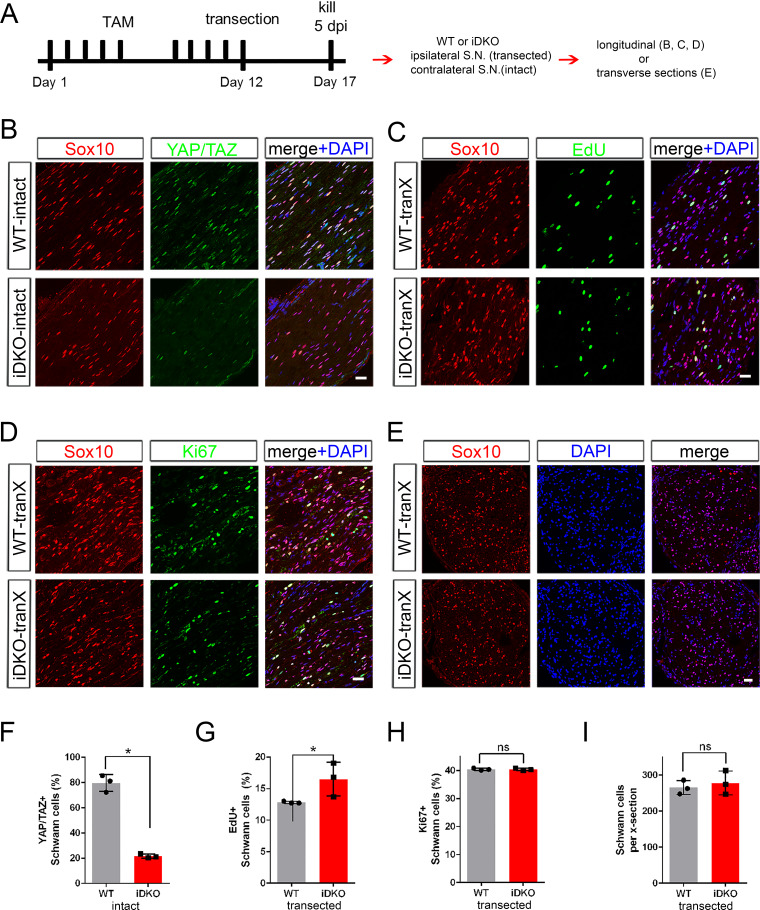
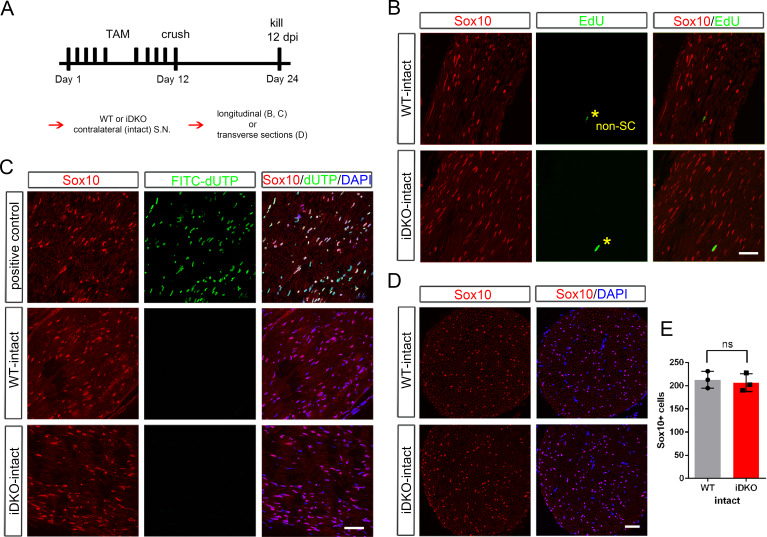
Figure 3. YAP/TAZ are dispensable for Schwann cell proliferation after axotomy.
(A) Schematic showing timeline of tamoxifen injection, sciatic nerve transection and sacrifice of adult WT or Yap/Taz iDKO. (B) Longitudinal sections of intact sciatic nerves showing efficient deletion of YAP/TAZ in iDKO. SC nuclei are marked by Sox10 (red). All cell nuclei are marked by DAPI (blue). (C) Longitudinal sections of transected nerves of WT or iDKO showing SCs in S-phase of the cell cycle marked by EdU (green). (D) Longitudinal sections of transected nerves of WT or iDKO showing proliferating SCs marked by Ki67 (green). (E) Transverse sections of transected nerves of WT or iDKO showing SCs marked by Sox10 (red). (F) Quantification of SCs expressing nuclear YAP/TAZ in intact sciatic nerves of WT or iDKO. n = 3 mice per genotype, *p=0.0495, Mann-Whitney. (G) Quantification of EdU+ SCs in transected nerves of WT or iDKO. n = 3 mice per genotype, *p=0.0463, Mann-Whitney. (H) Quantification of Ki67+ proliferating SCs in transected nerves of WT or iDKO. n = 3 mice per genotype, ns, not significant, p=0.5127, Mann-Whitney. (I) Quantification of Sox10+ SCs in transected nerves of WT or iDKO. n = 3 mice per genotype. ns, not significant, p=0.8273, Mann-Whitney. Scale bars = 30 μm (B–E).