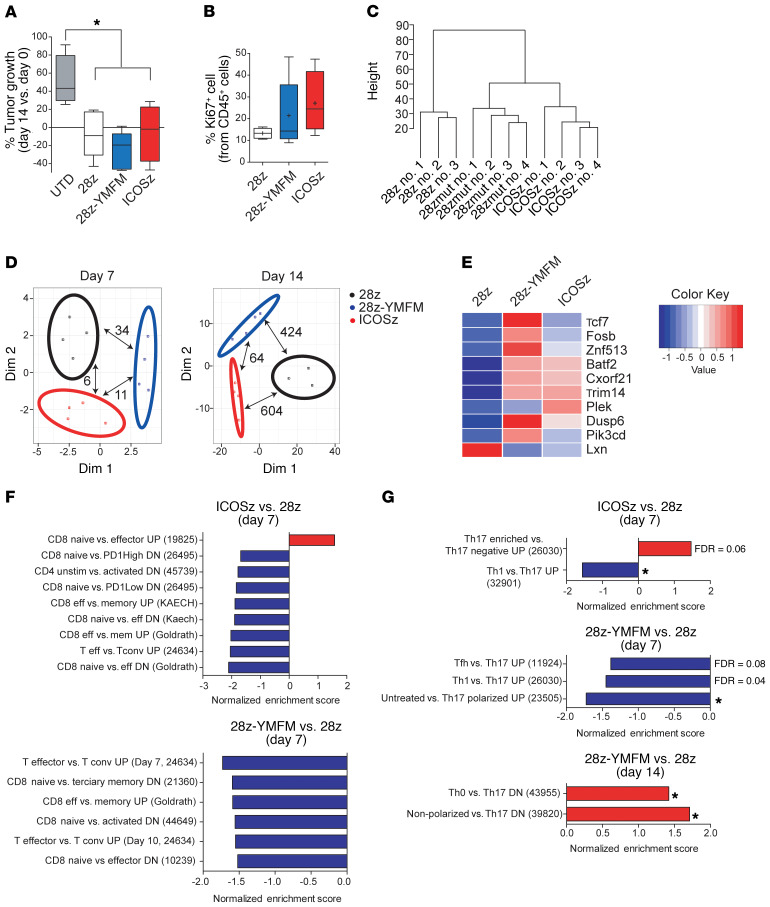
Figure 4. In vivo long-term signaling through 28z-YMFM CAR is associated with a transcriptional profile that resembles ICOS signaling.
NSG mice bearing s.c. Capan-2 tumors were treated with CAR-T cells and tumor growth was monitored. T cells were isolated from tumors on day 7 and 14 after treatment for analysis. (A) Box plots of the change in tumor volume on day 14 versus baseline (n = 4–5). *P < 0.05 by 1-way ANOVA with Tukey’s post hoc test. (B) The expression of CD45 and Ki67 on T cells isolated on day 14 after treatment was analyzed by flow cytometry. For A and B, box plots show median (line), mean (plus symbol), and 25th to 75th percentile (box). The end of the whiskers represents the minimum and the maximum of all of the data. Error bars represent ± SEM (n = 4–5). P > 0.05. (C) Dendrogram showing the relatedness of gene expression patterns for individual animals. (D) MDS analysis of differentially expressed genes between the 3 sets of CAR-T cells isolated from Capan-2 xenograft tumors (n = 3–4) on day 7 and day 14. The number of differentially expressed genes (FDR < 0.05, FC < 2 or FC > 2) between a pair of CARs is indicated with pair-connecting arrows. (E) Heatmap showing differential expression of genes associated with T cell differentiation and exhaustion. Genes reported to be preferentially expressed in early stages of T cell differentiation (Tcf7, Fosb, Znf513, Batf2, Cxorf2, Trim14) and genes upregulated in effector cells (Lxn) or downregulated in effector or exhausted T cells (Dusp6 and Pik3cd) are plotted. (F and G) Normalized enrichment scores of selected up- or downregulated gene sets associated with T cell differentiation (F) or Th17 polarization (G) as determined by GSEA using MSigDB C7 gene ontology sets. *P < 0.05. For all pathways, FDR q ≤ 0.02 (unless otherwise indicated). Reference numbers for immunology-related gene lists from MSigDB C7 are indicated in parentheses.