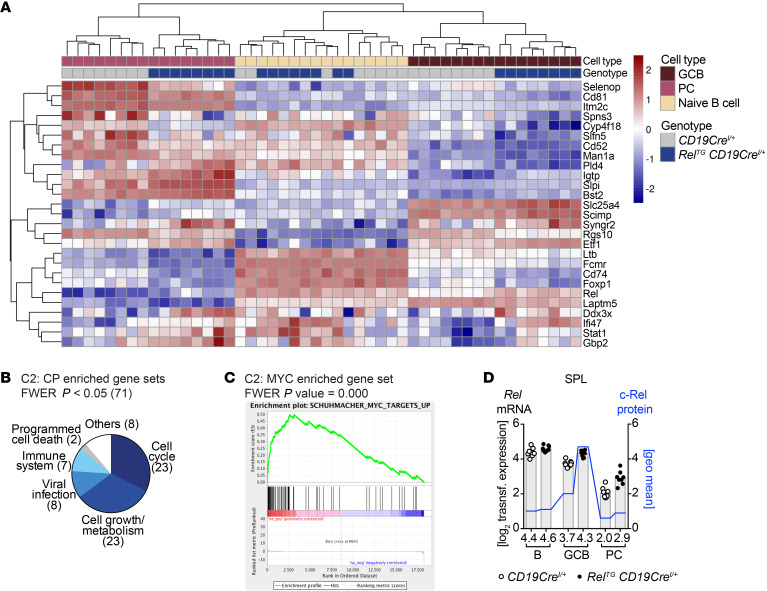
Figure 8. Strong enrichment of cell cycle and cell growth pathways in gene expression of c-Rel–overexpressing GCB cells.
(A) Common significantly differentially expressed genes (padj < 0.05) in GCB cells and plasma cells (PC) of RelTG CD19CreI/+ compared with control mice. Z-transformed expression values for each of 8 individual mice per genotype are shown in a heatmap representation. (B) Categories of significantly enriched gene sets in RelTG CD19CreI/+ GCB cell population evaluated by GSEA of C2: CP data set. (C) Enrichment plot of enriched gene sets in RelTG CD19CreI/+ GCB cell population with the highest normalized enrichment score among C2 gene sets containing the term MYC. (D) On the left x axis, log2 of transformed RNA expression values for Rel are plotted (gray bars). Individual data points are plotted. Bars and numbers below graphs are geometric means (n = 8). On the right x axis, geometric means of c-Rel protein levels are plotted (blue line) based on flow cytometry data presented in Figure 5A. FWER, family-wise error rate. See Supplemental Figure 10 and Supplemental Information.