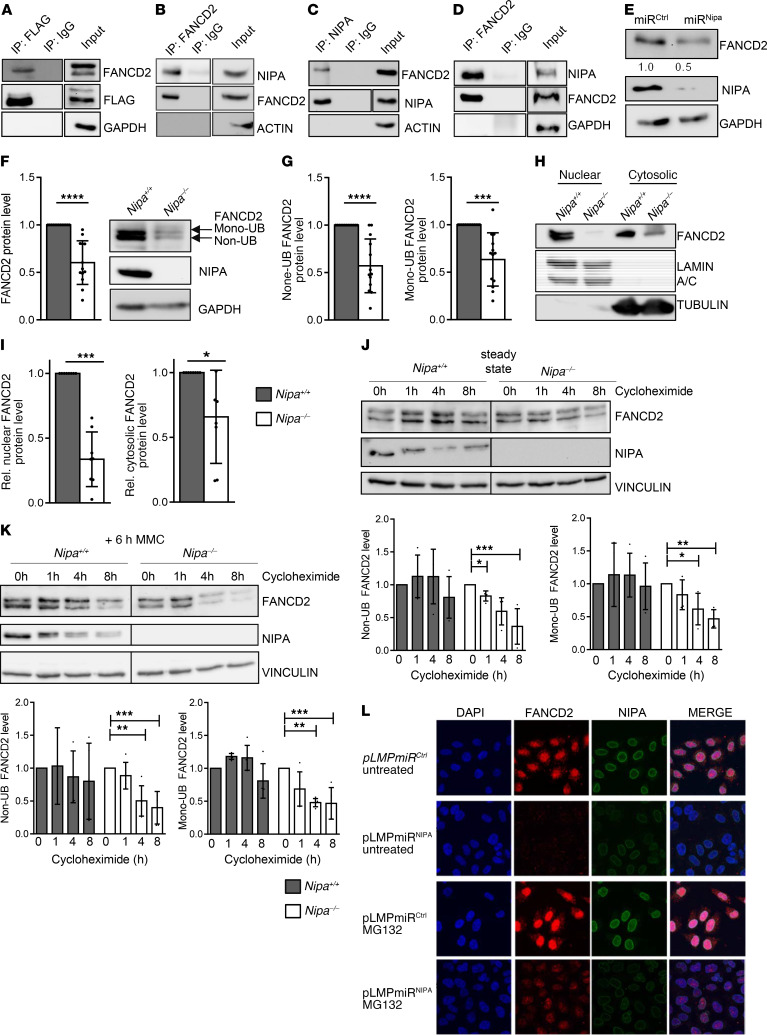
Figure 4. NIPA interacts with FANCD2.
(A) FLAG coimmunoprecipitation of Phoenix E cells transiently transfected with FLAG-hNIPA. (B) FANCD2 coimmunoprecipitation of Phoenix E cells transiently transfected with FLAG-hNIPA. (C) NIPA coimmunoprecipitation of HeLa cells with endogenous levels of NIPA. (D) FANCD2 coimmunoprecipitation of HeLa cells with endogenous levels of NIPA. (E) Western blot analysis of HeLa cells retrovirally transfected with pLMP miRCtrl or miRNipa for FANCD2, normalized to GAPDH. (F) Western blot analysis of Nipa+/+ and Nipa–/– primary MEFs for FANCD2, NIPA, and GAPDH. Quantification of relative FANCD2 protein levels normalized to GAPDH of Nipa+/+ and Nipa–/– primary MEFs. n = 13 Nipa+/+; n = 13 Nipa–/–. (G) Quantification of relative nonubiquitinated (Non-UB) and monoubiquitinated (Mono-UB) FANCD2 protein levels of Nipa+/+ and Nipa–/– primary MEFs analyzed by Western blot. n = 13 Nipa+/+; n = 13 Nipa–/–. (H) Western blot analysis of nuclear and cytosolic extracts of Nipa+/+ and Nipa–/– primary MEFs for FANCD2, lamin A/C, and tubulin. (I) Quantification of relative (rel.) FANCD2 protein levels normalized to lamin A/C (nuclear) or tubulin (cytosolic) of Nipa+/+ and Nipa–/– primary MEFs. n = 8 Nipa+/+; n = 8 Nipa–/–. (J) Western blot analysis of non- and mono-UB FANCD2 levels of Nipa+/+ and Nipa–/– primary MEFs (steady state) treated with cycloheximide for the indicated times. n =4 Nipa+/+; n = 4 Nipa–/–. (K) Western blot analysis of non- and mono-UB FANCD2 levels of Nipa+/+ and Nipa–/– primary MEFs (6 hours, 0.5 μM MMC) treated with cycloheximide for the indicated times. n = 5 Nipa+/+; n = 5 Nipa–/–. (L) Immunofluorescence for FANCD2, NIPA, and DAPI in untreated and 4-hour MG132–treated (5 μM) in HeLa cells retrovirally transfected with pLMP miRCtrl or miRNipa. Representative confocal microscopy images are shown. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001. A paired 2-tailed Student’s t test Original magnification, ×63. (F, G, and I) or Dunnett’s test (J and K) was used for statistical analyses. Data are presented as mean ± SD. See also Supplemental Figures 6–9.