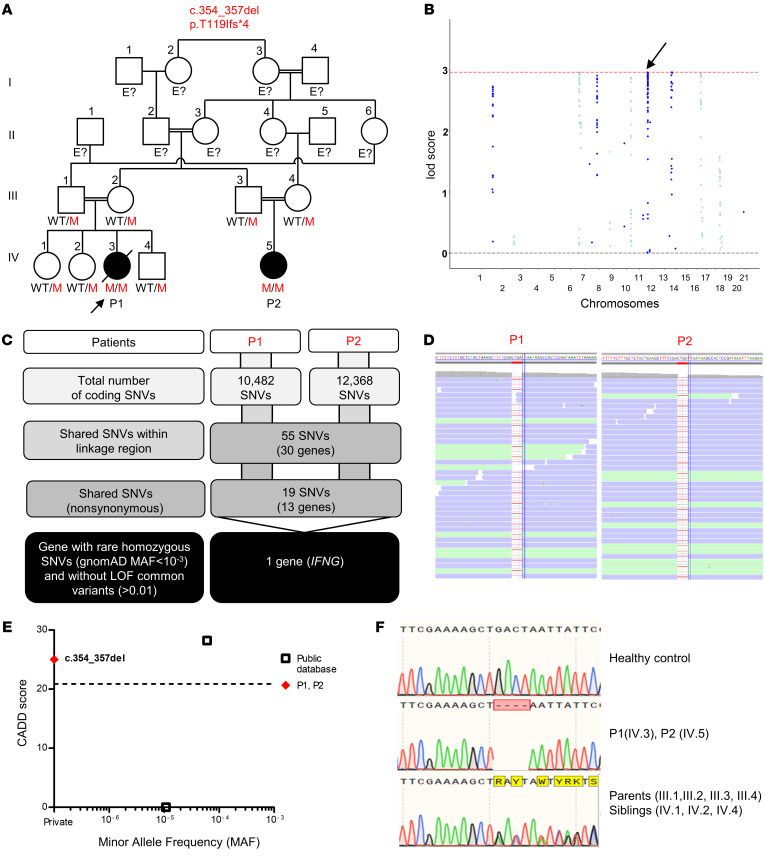
Figure 1. IFN-γ deficiency.
(A) Pedigree of 2 related kindreds, showing familial segregation of the c.354_357del (p.T119Ifs*4) mutant (M) IFNG allele. The affected patients are represented by black circles, and an arrow indicates the proband. Each generation is designated by a roman numeral (I–IV). E?, individuals of unknown genotype. (B) Linkage analysis. (C) An analysis of WES data identified IFNG as a candidate MSMD gene carrying a rare homozygous mutation common to P1 and P2 within the linkage regions. SNVs, single nucleotide variants. (D) Alamut viewer presentation of the region of the genome containing the mutation, in the sense orientation, for P1 and P2. (E) CADD score and MAF of all homozygous coding variants previously reported in public databases (gnomAD v2.1 and TOPMED/BRAVO) (http://gnomad.broadinstitute.org; https://bravo.sph.umich.edu) and in our in-house (HGID) database. The dotted line corresponds to the MSC with its 95% CI. The c.354_357del mutation is shown as a red diamond. (F) Sanger sequencing of the region containing the IFNG mutation from a healthy control, patients, and the patients’ heterozygous relatives.