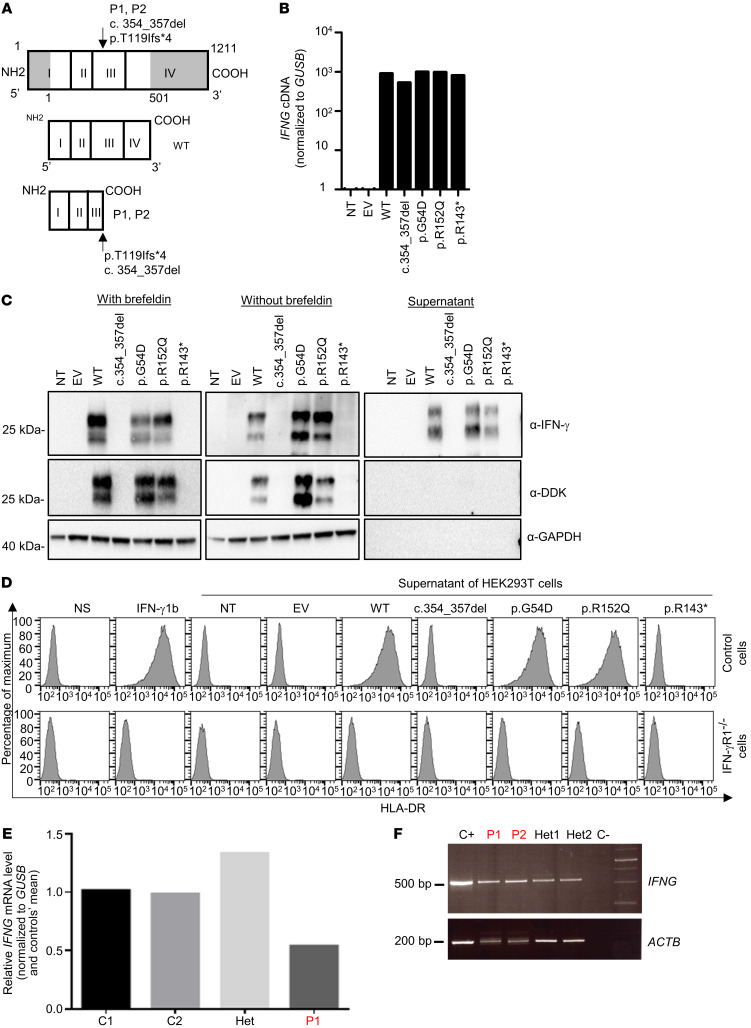
Figure 3. Levels of RNA and protein produced from the IFNG allele in an overexpression system and in vitro characterization.
(A) Schematic representation of the WT protein and predicted proteins for P1 and P2. (B) qPCR on cDNA from HEK293T cells nontransfected (NT) or transfected with an empty plasmid (EV), WT-IFNG, or mutated IFNG. GUSB was used for normalization. The results shown are representative of 2 independent experiments. (C) Western blot analysis of IFN-γ in HEK293T cells left NT or that were transfected with an EV, WT-IFNG, or mutated IFNG, all inserted into p.CMV6 with a C-terminal DDK tag, with (left) or without (middle) brefeldin treatment and the addition of supernatants from HEK293T-transfected cells (right). The anti–IFN-γ antibodies used were a monoclonal mouse anti-IFN-γ antibody recognizing an N-terminal epitope between amino acids 20 and 50, and an antibody directed against the C-terminal DDK tag. An antibody against GAPDH (α-GAPDH) was used as a protein-loading control. The results shown are representative of 2 independent experiments. Different exposure times were used for each Western blot. (D) Induction of HLA-DR on SV-40 fibroblasts from a healthy control and from a patient with AR complete IFN-γR1 deficiency. Cells were activated with commercial IFN-γ or supernatants obtained from HEK293T cells transfected with different constructs. The results shown are representative of 2 independent experiments. (E) qPCR on cDNA from the HVS-T cells from healthy travel controls (C1 and C2), a heterozygous individual, and P1. GUSB was used for normalization. The results shown are representative of 2 independent experiments. (F) RT-PCR of exons 1–4 of the IFNG cDNA in PHA blasts from a healthy control (C+), 2 patients (P1 and P2), their relatives (Het1 and Het2), and a negative control (C–). The ACTB gene was used as a cDNA loading control.