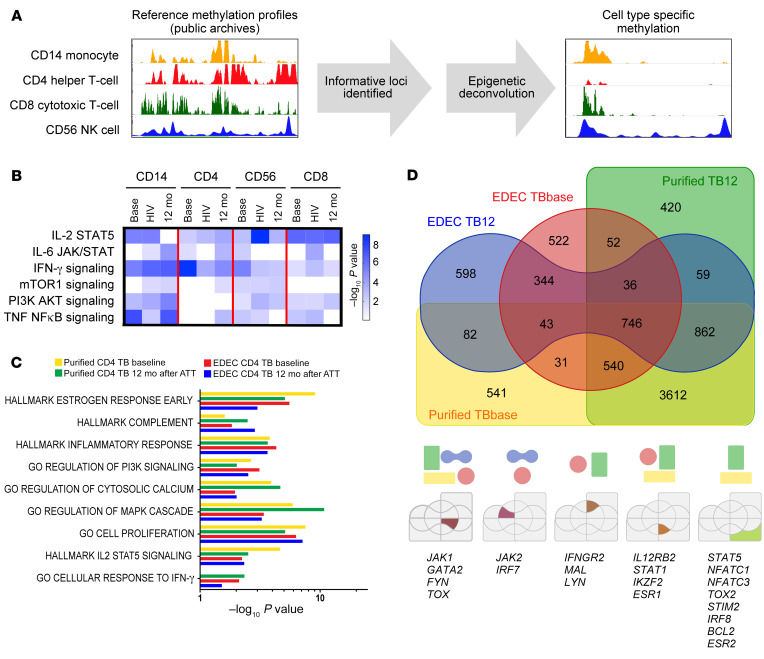
Figure 2. Global DNA methylation perturbations persist 6 months after successful ATT.
The Illumina DNA Methylation EPIC array was performed on bulk PBMCs from asymptomatic household contacts (n = 10) and individuals with TB (n = 15; TB/HIV– = 7; TB/HIV+ = 8) at baseline. All individuals with TB had treatment success. For individuals without HIV coinfection, DNA methylation status was evaluated at baseline and 12 months later, 6 months after completion of successful ATT. (A) To ascertain the immune cell–specific DNA methylation changes, cell-specific DNA methylation reference profiles were downloaded from public archives, informative loci were identified, and cell type–specific methylation profiles were identified. (B) Cell-specific DNA methylation differences are shown for TB patients (base) and TB/HIV (HIV) patients at baseline and for TB patients 12 months after baseline (12 mo), 6 months after completion of successful ATT. All results were compared with HC data. GO pathway analysis for monocytes (CD14+), Th cells (CD3–CD4+), NK cells (CD3–CD56+), and CTLs (CD3+CD8+). (C and D) Purified Th cells (CD3+CD4+) (n = 2) and EDEC-identified Th cells (CD3+CD4+) (n = 8 TB patients; n = 10 controls) were compared with cells from controls, and common hypermethylated pathways and genes were identified.