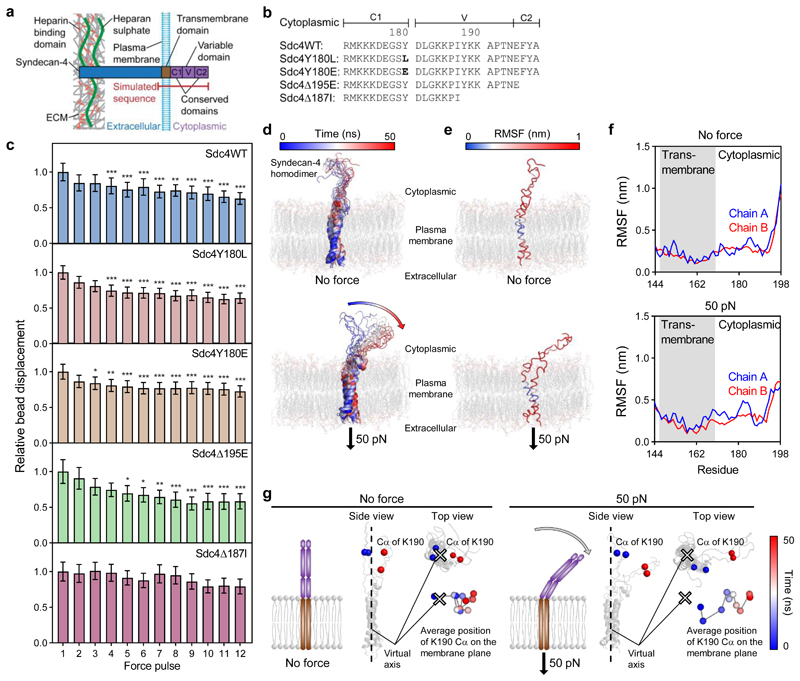
Figure 5. Syndecan-4 mechanotransduction requires the V-region which changes conformation under force.
a, Schematic representation of syndecan-4 structure. b, Cytoplasmic domain amino acid sequences for wild type syndecan-4 (Sdc4WT), syndecan-4 which cannot be phosphorylated on Y180 (Sdc4Y180L), phospho-mimetic syndecan-4 (Sdc4Y180E), syndecan-4 truncated in the C2 domain (Sdc4Δ195E) and syndecan-4 truncated in the V domain (Sdc4Δ187I). c, Displacement of syndecan-4 bound beads in response to pulsatile 1 nN force in mouse embryonic fibroblast cell lines expressing wild type or mutant syndecan-4 on fibronectin, relative to force pulse 1. See Supplementary Fig. 15 for single cell data. nSdc4WT = 27, nSdc4Y180L = 35, nSdc4Y180E = 43, nSdc4Δ195E = 23, nSdc4Δ187I = 22 cells; Friedman test with Dunn pairwise comparisons: *P ≤ 0.0238, **P ≤ 0.0092, ***P ≤ 0.001 vs force pulse 1. Mean ± s.e.m. d, Molecular structure snapshots of syndecan-4 at 5 ns intervals during a 50 ns simulation with no force applied (molecular dynamics (MD)) and with constant force pulling at 50 pN on the extracellular domain of syndecan-4 (steered molecular dynamics (SMD)). e-f, Root mean square fluctuation (RMSF) of syndecan-4 Cα atoms overlaid on molecular structure (e) and plotted against residue (f). g, Average position of lysine K190 Cα atoms on the membrane plane representing directed movement of the cytoplasmic domain from the virtual axis (located between transmembrane helices) towards the membrane in SMD. Average position was calculated for both K190 Cα atoms (chain A and B) in each 5 ns increment. See Supplementary Fig. 17 for amino acid sequence used in simulations.