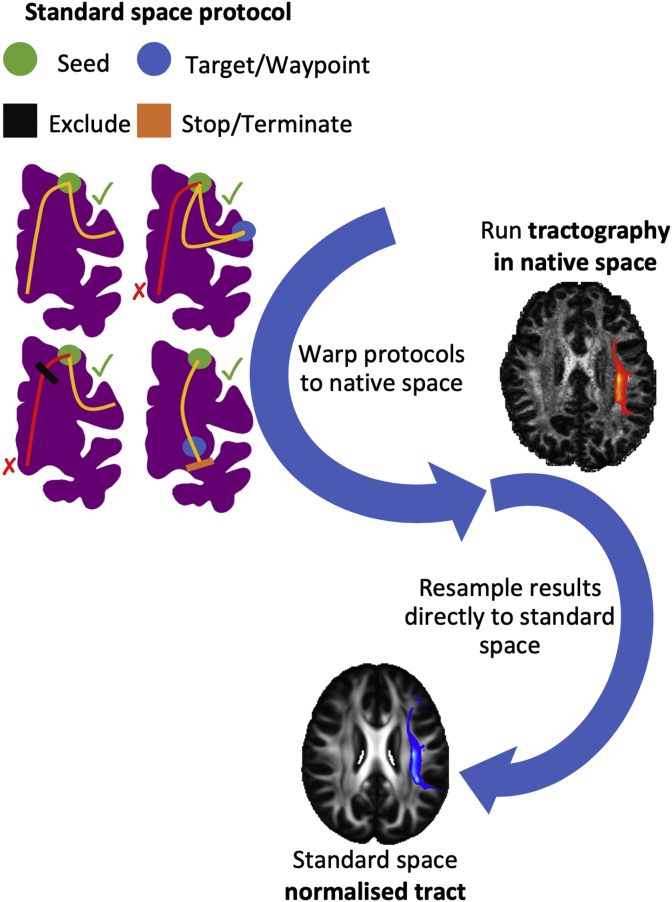
Fig. 1.
Schematic of the stages for automated tractography, as implemented in the “XTRACT” toolbox, with an example of the left arcuate fasciculus (AF) for the human brain. 1) Tractography protocol masks are defined in standard space with seed (green), exclusion (black), waypoint (blue) and termination (orange) masks (see the “Protocols” section for full details of definitions). 2) The protocol masks are warped to the subject’s native space using the subject-specific non-linear warp fields. 3) Probabilistic tractography is performed in the subject’s native space using the crossing fibre modelled diffusion data. Notice that tractography is performed in continuous coordinates in native diffusion space. Interpolation only occurs in the end when saving the result as a spatial histogram (i.e. tract visitation map). For this step nearest-neighbour interpolation is used at a user-specified spatial resolution. 4) The resultant tract stored in standard space, overlaid on the FSL_HCP1065 FA atlas.