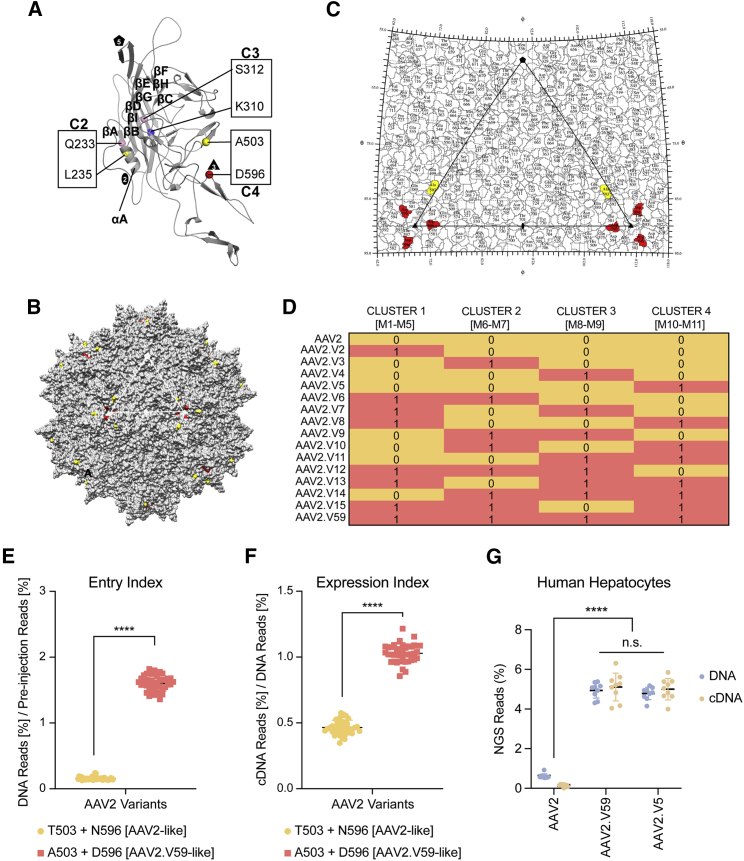
Figure 2.
Functional test of AAV2.V59 variants in humanised FRG mice.
(A) Structure and location of AAV2.V59 residues. Model of VP3 monomer colored gray. The residues in the VP3 clusters are shown as spheres, and the clusters are labeled as cluster 2 (C2), cluster 3 (C3), and cluster 4 (C4). (B and C) Surface map (B) and stereographic roadmap projection (C) of the 3D model viewed down the icosahedral 2-fold axis. The icosahedral 2-, 3-, and 5-fold axes, are depicted as an oval, a triangle, and a pentagon respectively. The non-polar residues L235 and A503 are colored yellow, polar residues Q233 and S312 are colored pink, basic residue K310 is colored blue, and acidic residue D596 is colored red. (D) Cluster composition of 16 AAV2 variants, including AAV2 and AAV2.V59. (See Figure S1 for composition of each cluster.) A yellow shadowed “0” indicates AAV2 origin for the whole cluster, whereas a shadowed salmon “1” indicates NP59 origin of the given cluster. (E and F) In vivo performance of the 16 AAV2 cluster variants in the humanized FRG (hFRG) model grouped by the origin of cluster 4 (AAV2 origin, yellow; NP59 origin, salmon). The results are shown as mean ± SD. (E) Entry index (2,014 vg/diploid human genome) and (F) the expression index. (G) In vivo comparison of AAV2, AAV2.V59, and AAV.V5 variants based on physical and functional transduction in the xenograft liver model. Percentage of NGS reads mapped to each capsid in human cells at the DNA (248 vg/diploid cell) and cDNA levels, normalized to the pre-injection mix, are shown. The results are shown as mean ± SD. Statistical significance was calculated using the two-tailed Mann-Whitney test. ∗∗∗∗p < 0.0001. n.s., not significant (p > 0.05).