Fig. 2.
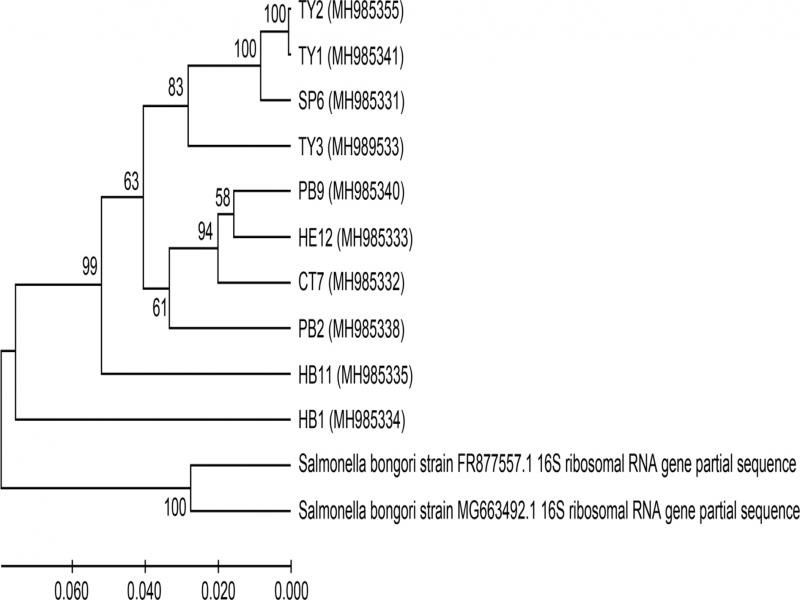
The phylogenetic tree constructed by MEGA X, with the 16S rRNA sequences of the 10 Salmonella strains and two out-groups of Salmonella bongori. The UPGMA method was used with 2000 bootstrap replications, bootstrap percentages are indicated on nodes. The ten Salmonella species in this study are denoted as previously in this study along with their NCBI accession numbers. Evolutionary distances were computed using the Maximum Composite Likelihood method and they are provided in the units of number of base substitutions per site. The rate variation among sites was modeled with a gamma distribution (shape parameter = 2). All positions containing gaps and missing data were eliminated. There were a total of 1034 positions in the final dataset.
