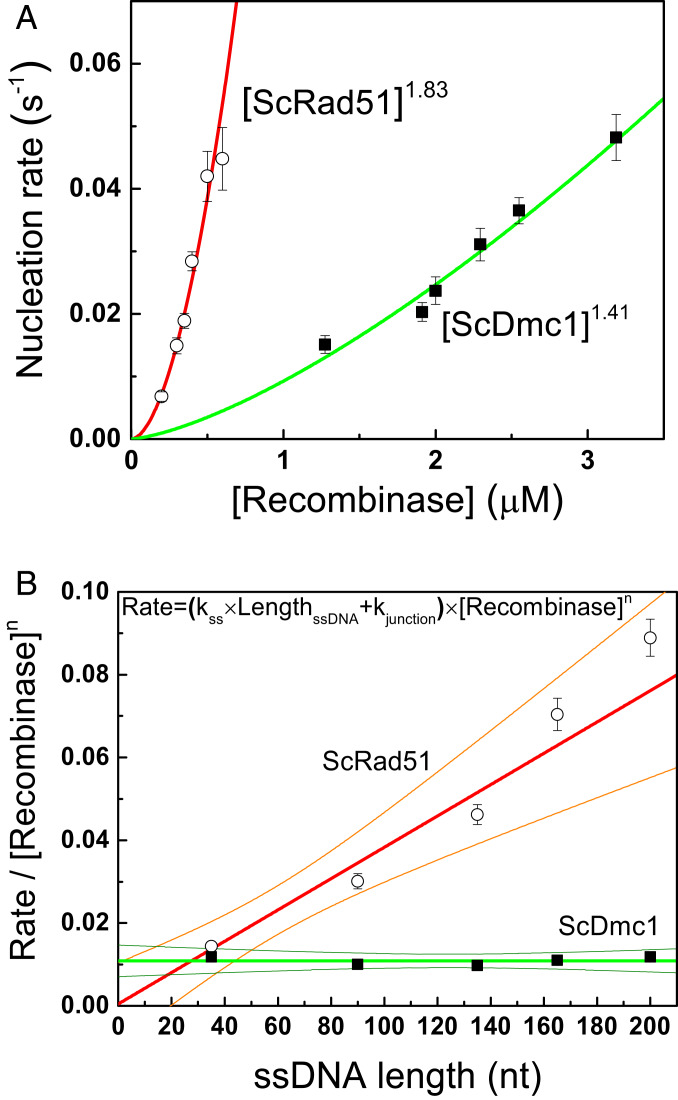
Fig. 3.
Dmc1 and Rad51 have the same nucleation unit during filament assembly, but different ssDNA affinities. (A) Nucleation rates of ScDmc1 and ScRad51 nucleoprotein filaments measured at different recombinase concentrations using 349/264 DNA substrates and 1 mM ATP. Similar power-law fittings of recombinase concentration return with n = 1.41 ± 0.17 for Dmc1 and n = 1.83 ± 0.17 for ScRad51. Nucleation times were determined from more than 50 assembled events of at least three independent experiments (SI Appendix, Fig. S3). The error bars of nucleation rate are the SD by bootstrapping 5,000 times. (B) Nucleation rates of ScDmc1 and ScRad51 have different dependences on ssDNA lengths of DNA substrates (351/dTn, n = 35, 90, 135, 165, and 200). Nucleation rates at each DNA length were determined at the 1.9-μM ScDmc1 or 0.4-μM ScRad51 (SI Appendix, Fig. S4). Apparent nucleation rates vary linearly with the ssDNA length, but with different slopes for ScDmc1 and ScRad51. The error bars of nucleation rate are the SD by bootstrapping 5,000 times.