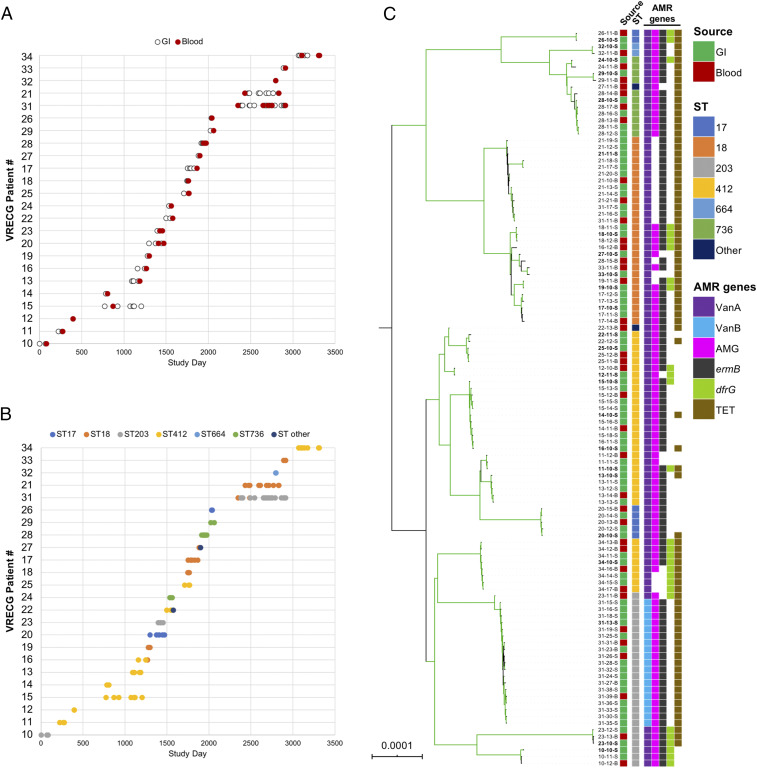
Fig. 1.
Sampling overview and population structure of 110 VREfm from immunocompromised pediatric patients. (A) Date of sampling of gastrointestinal (GI) (white) and bloodstream (Blood) (red) VREfm isolates from 24 study patients. Patients are labeled according to their “VRECG” patient number. (B) Distribution of STs for all isolates in the sample set. (C) Single-copy core genome phylogeny of all isolates in the sample set. Isolates are named as XX-YY-S/B, where XX is the patient number, YY is the isolate identifier, S indicates GI isolates, and B indicates blood isolates. The RAxML phylogeny is based on a gapless alignment of SNPs in the core genome identified by Snippy, with SNPs due to recombination removed by ClonalFrameML. Branches colored green are supported by bootstrap values >90. Tips are annotated with isolate names, source, ST, Van operon type, and other drug resistance-associated (AMR) genes identified in each genome. Isolate names in bold mark the patient-specific reference genomes used to identify variants in subsequent isolates in each patient. AMG, aminoglycoside-resistance genes; TET, tetracycline-resistance genes.