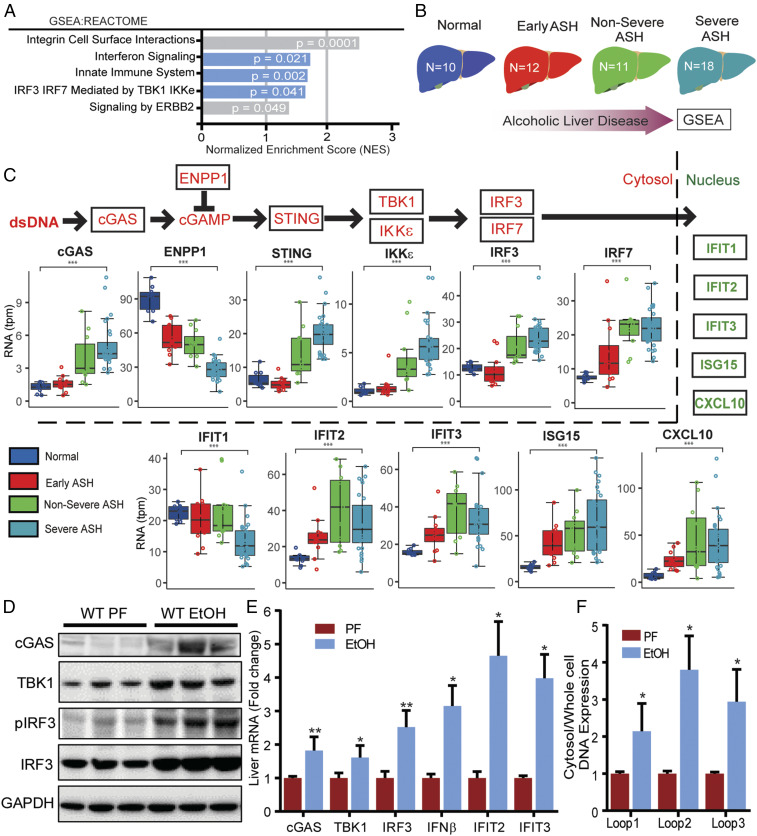
Fig. 1.
Alcohol activates the cGAS-IRF3 pathway in humans and mice. (A) GSEA of RNA-seq expression data from liver tissue of healthy controls and patients with alcohol-related liver disease (ALD). The enrichment score (ES) reflects the degree to which a gene set is overrepresented at the top or bottom of a ranked list of genes. (B) Schematic depicting the spectrum of disease severity within the ALD patient cohort. (C, Top) Abbreviated schematic of the cGAS-IRF3 pathway (red) and downstream IRF3-regulated genes (green). (C, Bottom) Differential expression analysis of cGAS-IRF3 pathway and IRF3-regulated genes based on disease severity within the ALD patient cohort. WT mice were fed 5% alcohol (EtOH) or a control PF diet for 6 wk. (D) Immunoblot for cGAS, TBK1, pIRF3, IRF3, and GAPDH and (E) hepatic mRNA expression of cGas, Tbk1, Irf3, and IRF3-regulated genes (Ifnβ, Ifit2, Ifit3) from EtOH and PF WT liver tissue. DNA in the cytosolic fraction hepatocytes from WT ETOH- and PF-fed mice was isolated, and the relative abundance of mtDNA was measured by RT-PCR normalized to whole-cell extract. (F) Cytosolic mtDNA content in liver tissue from WT and PF mice. N = 10 mice/group. Data are shown as mean ± SD. *P < 0.05; **P < 0.01; ***P < 0.001.