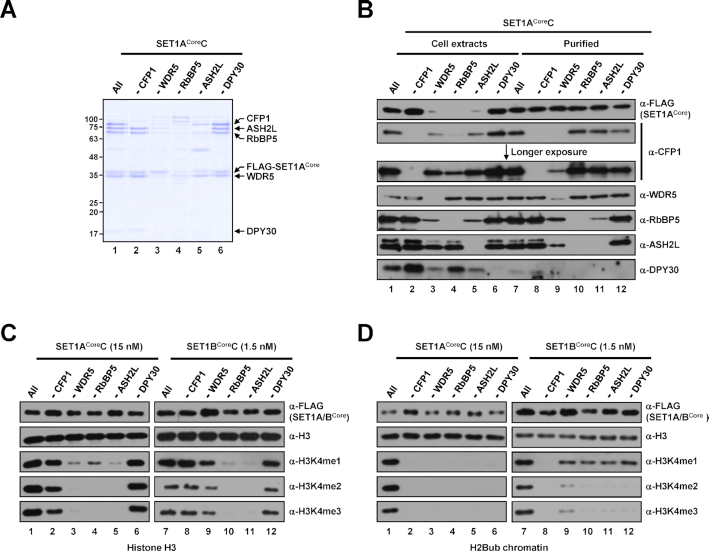
Figure 2.
Subunit requirements for H3K4 methylation activities of human SET1ACore and SET1BCore complexes. (A) SDS-PAGE/Coomassie blue staining of purified SET1ACore complexes reconstituted with baculoviruses in the absence of the indicated subunits. Sample loadings were normalized to FLAG-KMT2Core proteins in this and other figures. Note that because of inefficient complex formation (∼10–20-fold lower concentration compared with the intact complex), 5- and 2.5-fold lower amounts (maximum loading volumes) of RbBP5 and ASH2L-deficient complexes, respectively, relative to others were used for SDS-PAGE. (B) Immunoblot analyses of whole-cell extracts of Sf9 cells infected with baculoviruses in the absence of the indicated subunits and SET1ACore complexes purified from each cell extract. Note that equal volumes of cell extracts were loaded (lanes 1–6), and the loading of purified complexes was normalized to FLAG-SET1ACore protein (lanes 7–12). The slightly fast migrating band in anti-WDR5 immunoblot (lane 9) is considered an insect WDR5 homolog in this and other figures. The upper bands observed in anti-ASH2L immunoblot are considered a post-translationally modified form(s) of ASH2L in this and other figures. (C and D) H3 (C) and H2Bub chromatin (D) substrates were subjected to in vitro HMT assays with the indicated subunit-lacking SET1ACore (left) or SET1BCore (right) complexes. Note that because of large differences in intrinsic H3K4 methyltransferase activities (Figure 1), different concentrations of SET1ACore (15 nM) and SET1BCore complexes (1.5 nM) were used for assays.