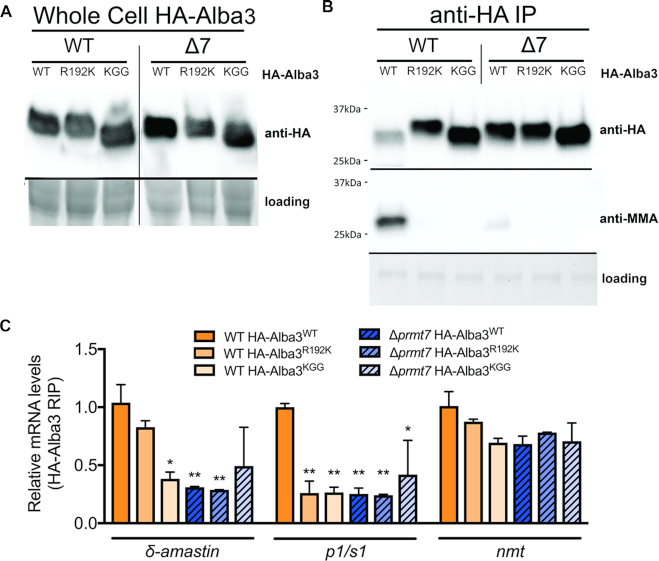
Figure 7.
Arginine methylation of Alba3 contributes to binding of δ-amastin and p1s1 mRNAs. (A) Endogenously tagged Alba3WT, RGG→KGG single point mutant (Alba3R192K) and hypomethylated mutant (Alba3KGG) showed no differential expression levels between WT and Δprmt7 cells. The migration of HA-Alba3KGG mutant protein is faster on SDS-PAGE, independent of PRMT7 levels. (B) Immunoprecipitation of HA-Alba3 shows arginine monomethylation is ablated in the RGG→KGG single point mutant (Alba3R192K), when all C-terminal RGG motifs are mutated into KGG (HA-Alba3KGG) and in Δprmt7 parasites. (C) Alba3 complexes were purified by RNA co-immunoprecipitation from in vivo UV-crosslinked cell lysates. Eluted RNA were analyzed by qRT-PCR and confirm that Alba3 binding to δ-amastin and p1/s1 nuclease decreases in the absence of PRMT7, but Alba3:nmt association remains constant independent of R mutation or PRMT7 levels. Interestingly, mutation of R192K disrupts Alba3:p1s1 association, but only mildly impacts Alba3:δ-amastin binding. Transcript levels are relative to 18S rRNA. *P< 0.05, **P< 0.01, Bonferroni two-way ANOVA test.