Figure 2.
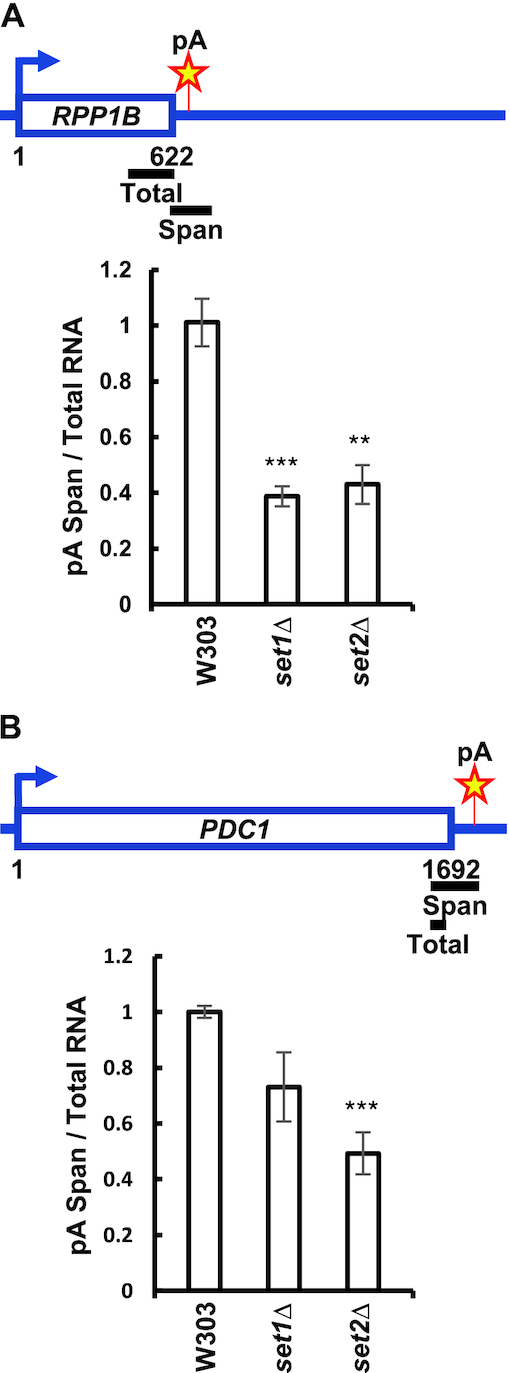
SET1 and SET2 deletion enhances utilization of single pA sites. (A, B) Schematic representation of primer pairs used for qRT-PCR analysis of transcripts reading through the PDC1 and RPP1B pA sites. Total RNA was reversely transcribed using random hexamers. qRT-PCR analysis of RNA was conducted using the primer pairs indicated above the bar graphs to determine the amount of total transcripts and that of transcripts spanning PDC1 and RPP1B pA sites, which represent unprocessed transcripts. The ratios of unprocessed to total transcripts in the mutant strains were normalized relative to the ratio in the wild-type W303 strain. Three biological replicates were performed for each gene. Bars show average values ± SD. **P < 0.01, ***P < 0.001 (Student's t-test).
