Figure 1.
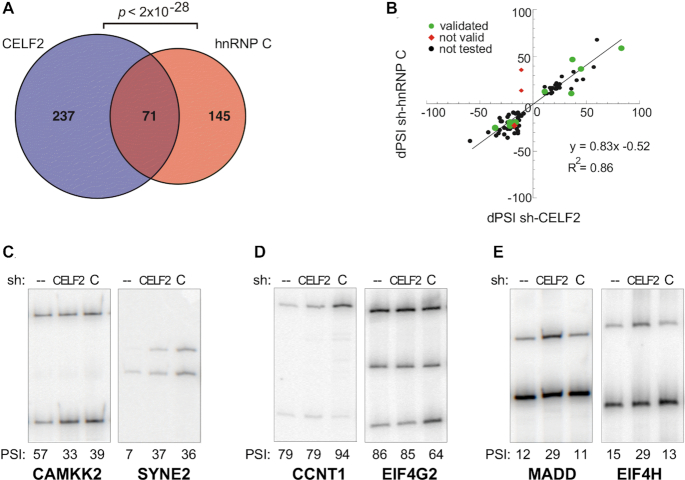
CELF2 and hnRNP C regulate a highly overlapping set of splicing events. (A) Overlap of events sensitive (|ΔPSI| > 10, P< 0.05) to either knockdown by shCELF2 or sh-hnRNPC in PMA stimulated Jurkat cells. Significance of overlap was calculated using a 2-tailed hypergeometric test. (B) Correlation of ΔPSI of the 71 splicing events sensitive to both shCELF2 and sh-hnRNP C in PMA stimulated Jurkat cells. Splicing events validated by RT-PCR are shown in green, while events that failed to validate by RT-PCR are shown as red diamonds. (C–E) Representative RT-PCR analysis of the splicing of genes predicted by RASL-Seq to be regulated by (C) both hnRNP C and CELF2, (D) hnRNP C only or (E) CELF2 only. Splicing is quantified by the percent of variable exon inclusion (PSI). Values and standard deviation from at least three biologically independent experiments is shown below in Figure 5.