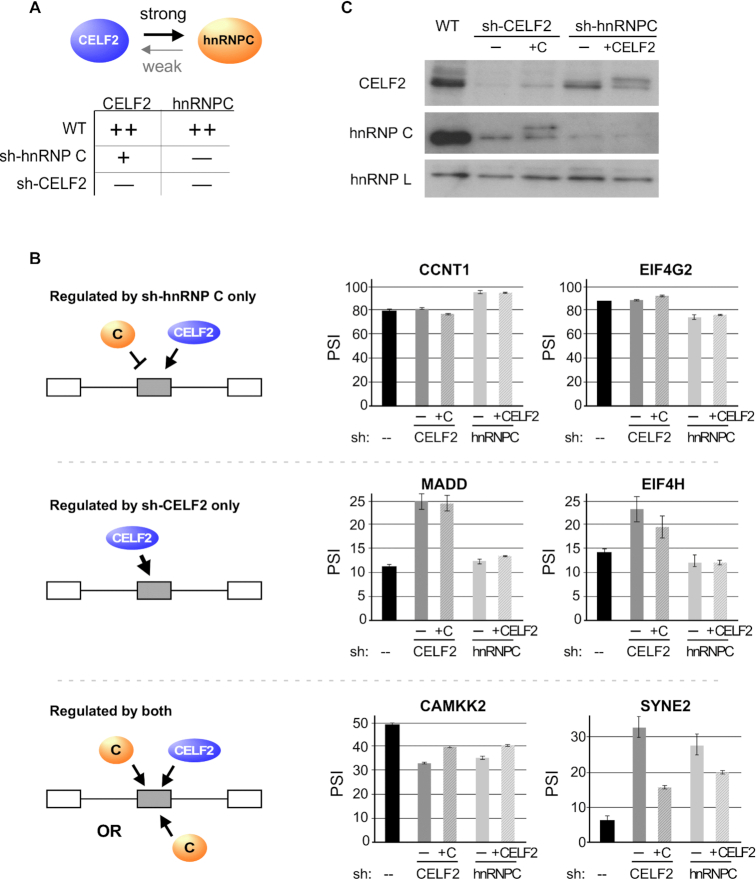
Figure 5.
Coordinated expression of CELF2 and hnRNP C fine-tune splicing outcomes of target genes. (A) Schematic and summary of the consequence of expression of shRNA against CELF2 (shC2) or hnRNP C (shC) on the expression of both proteins, as listed on the top of the columns. ++, + and − indicate high, medium and low expression. (B) Left: Potential models of action of CELF2 and hnRNP C on splicing targets that account for both the coordinated regulation observed between these proteins and the subset of target genes that are impacted by depletion of only one or the other protein. See main text for detailed description of models. Right: Quantification of RT-PCR analysis of several target genes upon partial rescue of CELF2 or hnRNP C expression in the knock-down of the other gene. Values are derived from at least three biologically independent experiments. Error bars represent standard deviation. (C) Western blot of CELF2 and hnRNP C expression in cells used for RT-PCR analysis in panel B. +C and +C2 indicate the overexpression of hnRNP C or CELF2, respectively, from Flag-tagged cDNA vectors. The mobility of the cDNA-expressed proteins is different from the endogenous due to the Flag tag. HnRNP L is used as a loading control.