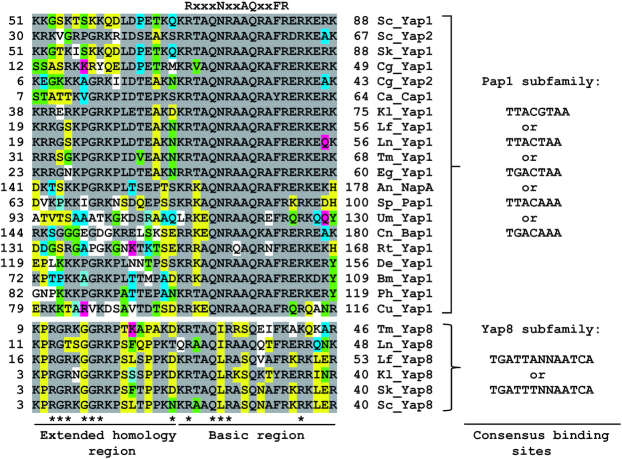
Figure 1.
Comparison of basic regions, the N-terminal adjacent sequences and consensus DNA binding motifs of Yap1/2 and Yap8 orthologues. The following proteins are from the Saccharomycotina (Ascomycota) species: Sc_Yap1 (NCBI accession no. NP_013707), Sc_Yap2 (NP_010711) and Sc_Yap8 (NP_015525) proteins are from S. cerevisiae, Sk_Yap1 (EJT43841) and Sk_Yap8 (EJT44313) are from S. kudriavzevii, Cg_Yap1 (XP_446996) and Cg_Yap2 (XP_446103) are from Candida glabrata, Ca_Cap1 (EEQ44283) is from C. albicans, Kl_Yap1 (CAH02665) and Kl_Yap8 (CAG99045) are from Kluyveromyces lactis, Lf_Yap1 (SCW02819) and Lf_Yap8 (SCW01455) are from Lachancea fermentati, Ln_Yap1 (SCU88970) and Ln_Yap8 (SCV05062) are from L. nothofagi, Tm_Yap1 (TOMI0S02e09538g1, Genome Resources for Yeast Chromosomes, http://gryc.inra.fr) and Tm_Yap8 (TOMI0S05e00210g1) are from Torulaspora microellipsoides, e.g._Yap1 (NP_984291) is from Eremothecium gossipii. An_NapA (XP_680782) is from Aspergillus nidulans (Pezizomycotina, Ascomycota). Sp_Pap1 (NP_593662) is from Schizosaccharomyces pombe (Taphrinomycotina, Ascomycota). Cn_Bap1 (XP_012046219) is from Cryptococcus neoformans (Agaricomycotina, Basidiomycota). Um_Yap1 (KIS70678) is from Ustilago maydis (Ustilaginomycotina, Basidiomycota). Rt_Yap1 (CEE11106) is from Rhodotorula toruloides (Pucciniomycotina, Basidiomycota). De_Yap1 (RHZ80237) is from Diversispora epigaea (Mucormycota). Br_Yap1 (ORY02218) Basidiobolus meristosporus (Zoopagomycota). Ph_Yap1 (TPX58997) is from Powellomyces hirtus (Chytridiomycota). Cu_Yap1 (ORZ35932) is from Catenaria anguillulae (Blastocladiomycota). Conserved amino acid residues involved in direct binding to DNA bases as determined for the S. pombe Pap1 protein (2) are indicated at the top of sequence alignment. Known residues that are important for Yap8 function are marked with asterisks (18, this work). Identical or similar amino acid residues are highlighted accordingly. Experimentally confirmed consensus DNA binding motifs for each subfamily are indicated on the right panel.