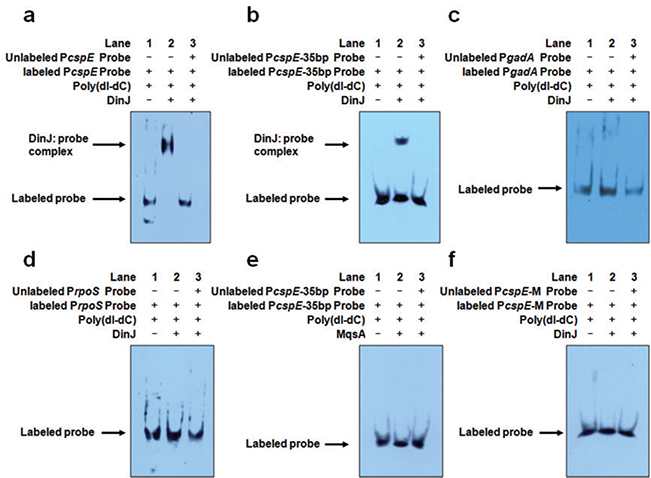
Fig. 3.
DinJ binds PcspE at the LexA palindrome but does not bind PrpoS.
A. The 176 bp DNA fragment of the cspE promoter (PcspE) was incubated with DinJ. Lanes 1–3: 6 ng of biotin-labelled PcspE; Lane 2: addition of 1.2 μg DinJ; and Lane 3: addition of 1.2 μg DinJ and 600 ng of unlabelled PcspE.
B. The 35 bp DNA fragment of cspE promoter (PcspE-35bp) was incubated with DinJ. Lanes 1–3: 30 pg of biotin-labelled PcspE; Lane 2: addition of 1.2 μg DinJ; and Lane 3: addition of 1.2 μg DinJ and 12 ng of unlabelled PcspE.
C. The 185 bp DNA fragment of gadA promoter (PgadA) was incubated with DinJ as a negative control. Lanes 1–3: 6 ng of biotin-labelled PgadA; Lane 2: addition of 1.2 μg DinJ; and Lane 3: addition of 1.2 μg DinJ and 600 ng of unlabelled PgadA.
D. The 185 bp DNA fragment of rpoS promoter (PrpoS) was incubated with DinJ. Lanes 1 to 3: 6 ng of biotin-labelled PrpoS; Lane 2: addition of 1.2 μg DinJ; and Lane 3: addition of 1.2 μg DinJ and 600 ng unlabelled PrpoS.
E. The 35 bp DNA fragment of cspE promoter (PcspE-35bp) was incubated with MqsA as a negative control. Lanes 1–3: 30 pg of biotin-labelled PcspE; Lane 2: addition of 1.2 μg MqsA; and Lane 3: addition of 1.2 μg MqsA and 12 ng of unlabelled PcspE. F. The mutated 35 bp cspE promoter (PcspE-M) lacking the LexA palindrome was incubated with DinJ. Lanes 1–3: 30 pg of biotin-labelled mutated PcspE; Lane 2: addition of 1.2 μg DinJ; and Lane 3: addition of 1.2 μg DinJ and 12 ng of unlabelled mutated PcspE. The unspecific competitor poly(dI-dC) was used for all samples.