Figure 1: Workflow of HMI-PRED.
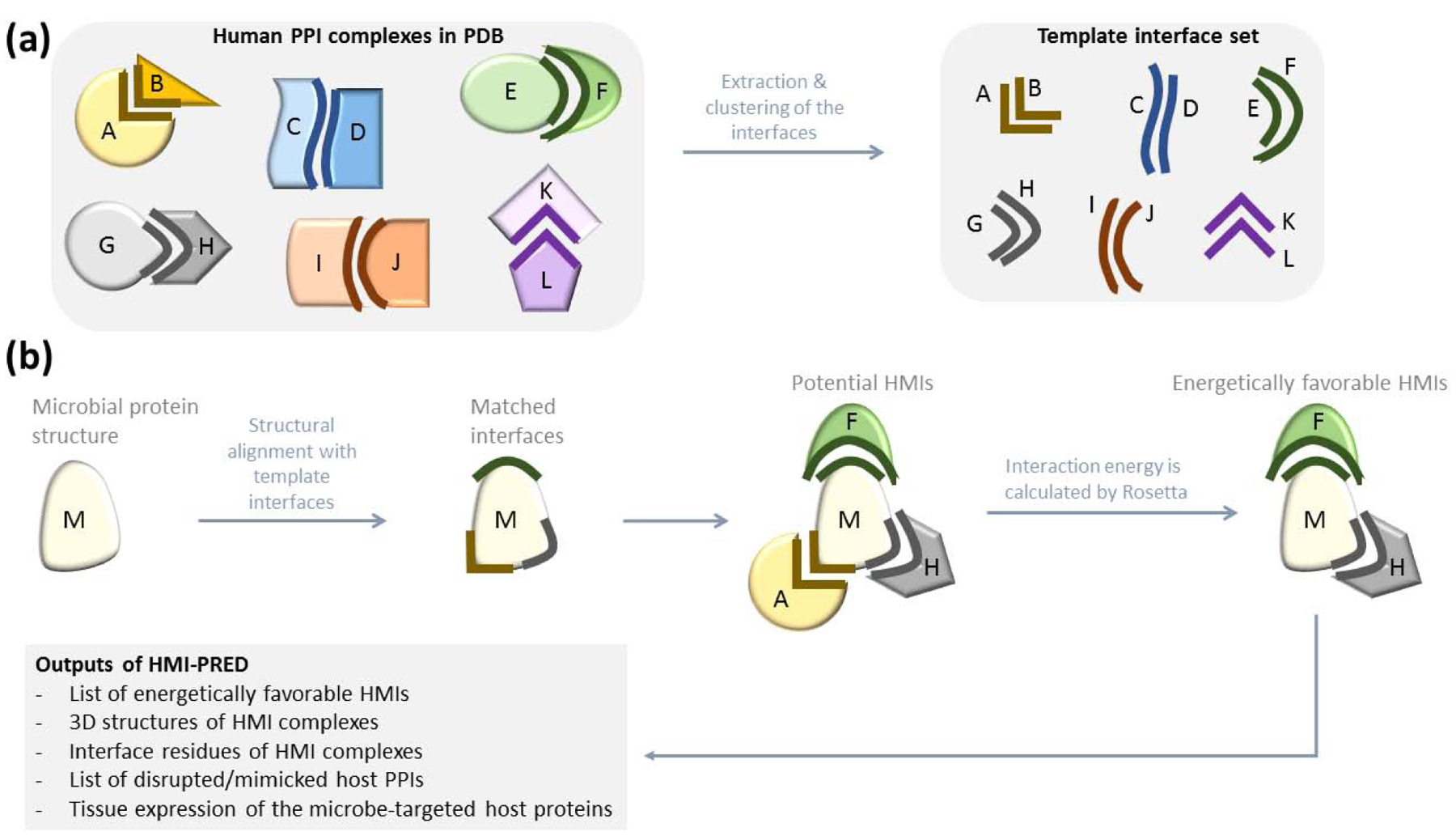
(a) Preparation of template interface set. We extract all human interfaces from PDB. Each interface has two faces and at least one face is from human. If both faces are from human, it is an endogenous interface. If one face is from human and the other face from another organism, it is an exogenous interface. Our non-redundant template interface set has 15.762 endogenous and 1.589 exogenous interfaces. (b) HMI-PRED workflow. Microbial protein structure is the only user input to HMI-PRED. If it has not been resolved yet, homology model can also be used. We first extract the surface of the microbial protein, then structurally align each template interface with the microbial protein surface and find some matches. If the microbial protein is aligned with the B-face of the interface, we assume that it can interact with the complementary face, A. We generate the potential HMI pairs based on this assumption. However, structural complementarity may not always guarantee electrochemical complementarity. Therefore, we calculate the interaction energy of the potential HMIs to pick the energetically favorable ones.
