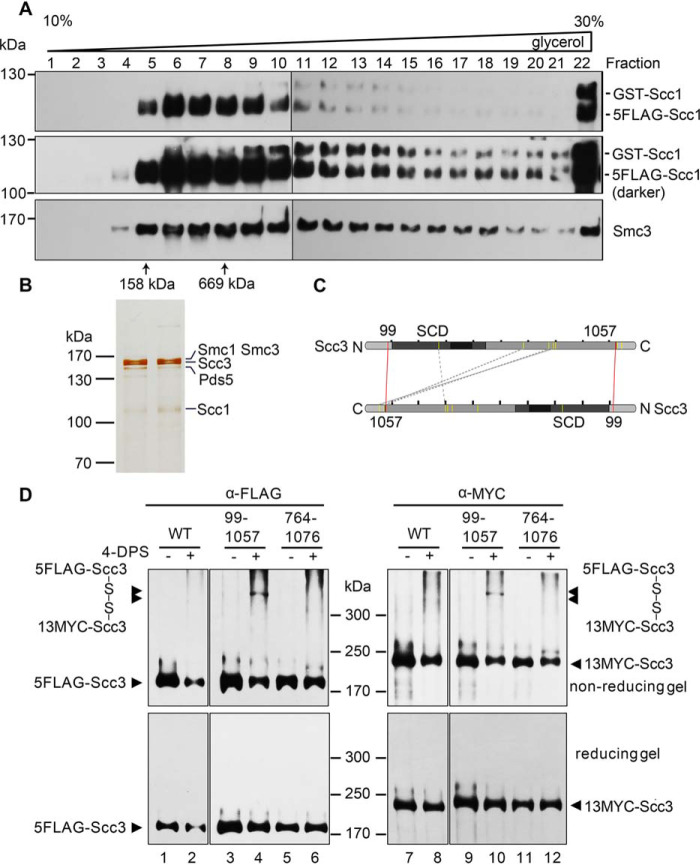
Figure 2.
Purification and cross-linking of the cohesin complexes. A and B, purification of the native cohesin complexes. The cohesin complexes were isolated from the yeast cells expressing p5FLAG-SCC1 cells via one-step affinity purification (i.e. anti-FLAG M2 column chromatography and FLAG peptide elution) followed by 10–30% glycerol density gradient centrifugation. The sample was divided into 24 fractions (0.5 ml each). After separation by SDS-PAGE, they were analyzed by IB with the indicated antibodies (A) or silver staining (B). The sedimentation of standard proteins (158 and 669 kDa) is indicated by an arrow. The band of each subunit was validated by MS as well. C, an Scc3-Scc3 connectivity map of the high confidence DSS cross-links detected in CXMS. The purified Scc3-containing complexes were cross-linked by DSS prior to trypsin digestion and LCMS/MS as described under “Experimental procedures.” The cross-linked amino acids were identified using the pLink search engines and labeled by a dashed gray line. The pair of amino acids validated by the following cysteine substitution and in vivo cross-linking are labeled by a red line. D, cysteine substitution of Lys-99 and Lys-1057 at the putative Scc3-Scc3 interface supports the in vivo cross-linking. Cys screening of the putative pairs near the intermolecular interface of Scc3 was conducted. The indicated pairs of amino acid residues (e.g. Lys-99/Lys-1057 and Lys-764/Lys-1076) in two copies of Scc3 were substituted by cysteine. WT or cysteine-substituted mutant cells were grown and treated with 180 μm 4-DPS (+) or DMSO (−) before harvest. The proteins were extracted and analyzed by nonreducing (top) or reducing (bottom) SDS-PAGE followed by anti-FLAG and anti-MYC IBs. The monomeric and dimeric Scc3 are indicated by single and double arrows, respectively.