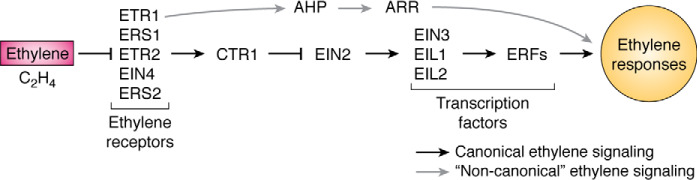
Figure 1.
Simple genetic model of ethylene signaling. In black is shown a model for ethylene signaling based on molecular genetic experiments in Arabidopsis. These experiments showed that ethylene signaling involves ethylene receptors (ETR1, ERS1, ETR2, EIN4, and ERS2), the protein kinase CTR1, and EIN2 that signals to the transcription factors EIN3, EIL1, and EIL2. These, in turn, signal to other transcription factors, such as the ERFs, leading to ethylene responses. This has long been considered the canonical signaling pathway. In this model, CTR1 is a negative regulator of signaling. Ethylene functions as an inverse agonist, where it inhibits the receptors, which leads to lower activity of CTR1 releasing downstream components from inhibition by CTR1. More recent evidence has shown the existence of an alternative, “noncanonical” pathway (in gray), where ETR1 signals to histidine-containing AHPs and then to ARRs to modulate responses to ethylene.