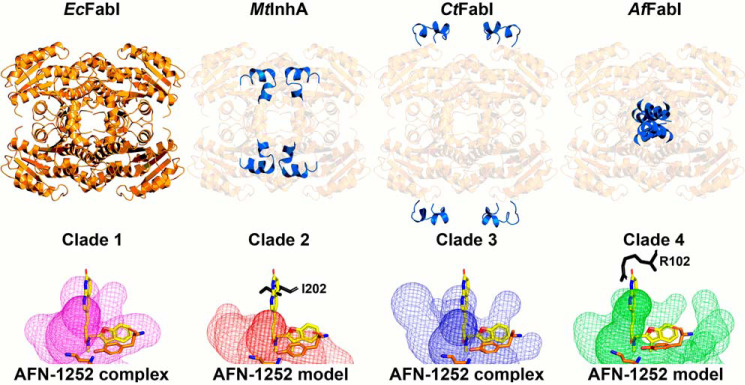
Figure 10.
Unique structural features of the FabI clades. Representative models for the four FabI clades are compared using clade 1 FabI as the standard for comparison. Top panel, clade 1 EcFabI has the basic Rossmann fold and tetrameric organization common to all FabIs. The unique features in clades 2–4 are highlighted in blue. Clade 2 enzymes are represented by the Mycobacterial enoyl-ACP reductase (MtInhA). Clade 2 enzymes contain an extended active site lid to accommodate the long acyl-ACP substrates (28 carbons) that arise during the synthesis of mycolic acids. Clade 3 plastid enzymes are represented by the Chlamydial CtFabI and contain peripheral loops of unknown function. Clade 4 enzymes are presented by AfFabI and contain carboxyl-terminal α9–α9 coiled coils that are required for high-affinity NADH binding. Bottom panel, active site volumes for each clade are depicted by a surface mesh. The catalytic lysine and tyrosine residues are shown in orange. Ternary complex structures of clade 1 (PDB ID: 4JQC) (77) and 3 (PDB ID: 4Q9N) (29, 83) enzymes with NAD(H) and AFN-1252 (shown in yellow) have been determined and contain active site volumes that bind AFN-1252. Binary complex structure of clades 2 (PDB ID: 4DRE) (78, 82) and 4 (PDB ID: 6VLY) enzymes with NAD(H) have been determined, and AFN-1252 was modeled in their active site volumes with respect to the tyrosine. In both cases, the 3-methylbenzofuran moiety of AFN-1252 extends outside the active site and the oxotetrahydronaphthyridine moiety clashes with amino acid residues (shown in black).