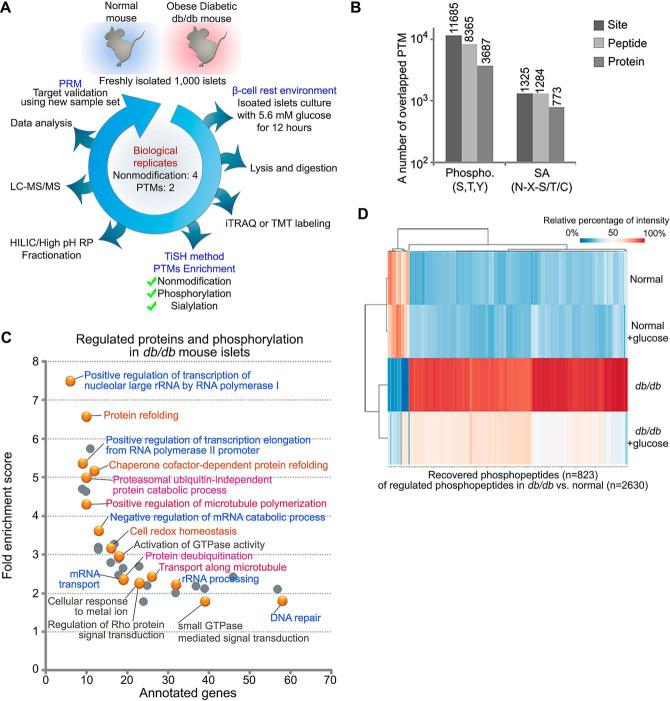
Fig. 1.
Quantitative global phosphoproteomic study. A, islets were isolated from db/db mice and WT mice and incubated with or without euglycemia (5.6 mm, 12 h) after which islets lysates were prepared and digested with trypsin/LysC. Tryptic peptide samples from individual experiments were labeled with isobaric mass tags (iTRAQ/TMT) and the peptides from equal amounts of independent experiments, then phosphopeptides and formerly sialylated N-linked glycopeptides (SA) were enriched by TiSH method. Each sample from modification groups were fractionated using HILIC or high pH reverse phase before being analyzed by LC-MS/MS. Twenty-five differentially regulated peptides were validated using Parallel Reaction Monitoring (PRM) assay with heavy isotope-labeled synthetic peptide. B, Histograms show the overlapping 11,685 and 1325 posttranslationally modified site, 8365 and 1284 peptides, and 3687 and 773 proteins in the phosphorylation group and the SA group, respectively. C, Heatmap of the recovered phosphopeptides (opposite to D/N; at least log2 fold change ± 0.3 in DG/D) ordered by hierarchical clustering. D: db/db mouse islets, N: wild type mouse islets, DG: db/db mouse islets exposed to euglycemia. Values for each phosphopeptide (column) at all analyzed samples (row) are color code based on the relative percentage of normalized intensity, low (blue) and high (red). D, Enriched biological processes for significantly regulated proteins with phosphorylation that exhibited restoration. Panther Gene Ontology enrichment analysis showing the fold enrichment score for the indicated dots (y axis) as well as the number of proteins assigned to classified functions (x axis). Corrected p value < 0.05.