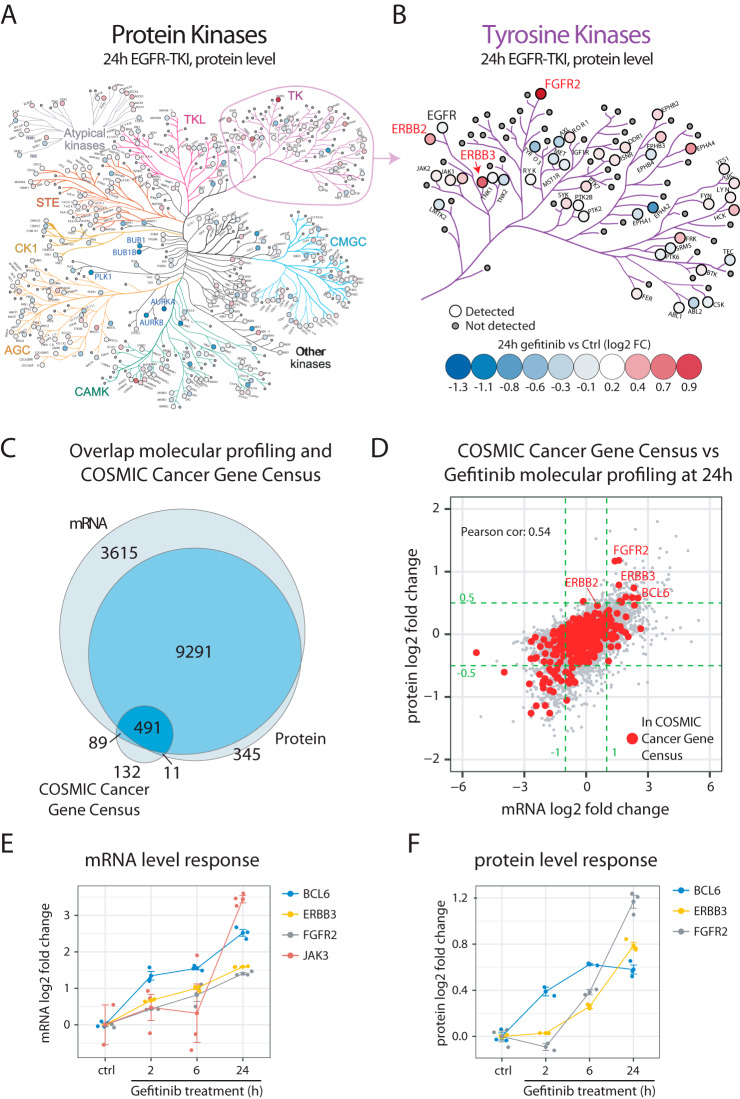
Fig. 2.
EGFR-TKI treatment results in upregulation of proteins potentially involved in treatment escape. A, Kinase regulation in response to EGFR-TKI treatment (gefitinib 2.5 μm, 24h) of A431 cells as measured at protein level. The map shows all kinases as visualized by the KinoViewer tool (30). Indicated in the map is also the fold regulation (blue-red scale). B, Zoom in on tyrosine kinases. Indicated in the map is EGFR (unchanged) as well as a few upregulated receptor tyrosine kinases with potential impact on therapy response (ERBB2, ERBB3 and FGFR2). C, Venn diagram indicating the overlap between the molecular response profiling and the COSMIC cancer gene census catalogue of genes that are causally implicated in cancer (43). D, Scatterplot showing the mRNA and protein level regulation at 24 h post treatment with gefitinib. Indicated in red are the COSMIC cancer gene census genes. Green dotted lines indicate the cutoffs used to define regulated mRNAs (log2 FC>±1), and proteins (log2 FC>±0.5). Indicated in the plot are a few upregulated cancer-associated genes potentially involved in EGFR-TKI treatment escape. E, mRNA level response to EGFR-inhibition by gefitinib shown for BCL6, ERBB3, FGFR2 and JAK3. F, Protein level response to EGFR-inhibition by gefitinib shown for BCL6, ERBB3 and FGFR2.