Figure 7.
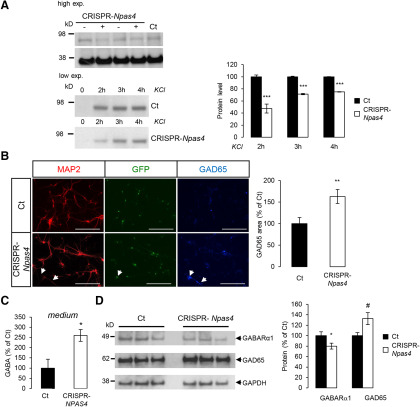
NPAS4 silencing by CRISPR-Cas9 mimics cell phenotype observed in APP-deficient neurons. Changes on inhibitory (GABA) synapses was analyzed after NPAS4 silencing A, left panel, Cortical neurons infected with CRISPR-Cas9 lentivirus targeting NPAS4 gene (CRISPR-NPAS4) show reduced NPAS4 levels as measured by Western blotting (high exposure). Same experiments were conducted after membrane depolarization with 50 mm KCl. NPAS4 accumulations was detectable by Western blotting at low exposure. Viruses without sgRNA were used as controls (Ct). Right panel, Quantification of NPAS4 protein level after 2, 3, and 4 h of KCl depolarization. Results (mean ± SEM) are expressed as percentage of non-treated controls Ct (N = 2); ***p < 0.0001 Student’s t test. B, Cortical neurons infected with CRISPR-NPAS4 lentiviruses at DIV1 were immunostained against MAP2 and GAD65 at DIV7. Quantification of GAD65 signal was normalized to the number of cells (five fields per coverslip, two coverslips for each genotype in two independent experiments (N = 2). Results (mean ± SEM) are given as percentage of control (Ct). Scale bar: 200 µm; **p = 0.0024. Mann–Whitney test. C, Quantification of GABA in culture medium at DIV7 of infected control neurons (Ct) and CRISPR-NPAS4-infected neurons. Results (mean ± SEM) are expressed as percentage of Ct (n = 5, N = 2); *p = 0.0146, Student’s t test. D, Neurons harvested at DIV7 and cell extracts analyzed by Western blotting for GABARα1, GAD65, and GADPH expression. Quantification of GABARα1 and GAD65 were normalized to GAPDH expression. Results (mean ± SEM) are expressed as percentage of Ct (n = 8, N = 3); *p = 0.049, #p = 0.0247, Student’s t test.