Abstract
Direct acting antivirals (DAAs) have revolutionized hepatitis C virus (HCV) treatment, but drug resistance could undermine proposed global elimination targets. Real‐world studies are needed to inform the impact of widespread DAA treatment on antiviral resistance in the community. The prevalence and range of posttreatment resistance‐associated substitutions (RASs) was determined in Australian patients with open access to DAAs through a wide range of prescribers. NS3, NS5A, and NS5B regions were amplified by polymerase chain reaction and analyzed by population sequencing. Clinically relevant RASs were identified using online databases (ReCALL and Geno2Pheno[hcv]). Of 572 samples, 60% were from genotype 3 and 27% from genotype 1a. Ninety‐two percent of people failed a DAA regimen containing an NS5A inhibitor, including 10% with a pangenotype regimen. NS5A RASs were detected in 72% of people with genotype 1 and 80% with genotype 3. For genotype 1, there was a range of RASs across the NS5A region, while for genotype 3, the Y93H RAS predominated (72%). The prevalence of NS3 RASs was higher in people exposed to an NS3 inhibitor (35% vs. 3.9%; P < 0.0001). NS5B resistance was rare, with a single case of sofosbuvir resistance. Multiclass drug resistance was found in 33% of people exposed to both NS3 and NS5A inhibitors. Conclusion: The high prevalence of NS5A RASs among people failing DAA therapy reinforces the importance of specific retreatment regimens, ideally guided by resistance testing. The impact of multiclass drug resistance on retreatment in people exposed to both NS3 and NS5A inhibitors needs to be assessed in real‐world studies. Surveillance for increasing antiviral resistance during treatment scale‐up is essential to maintain the efficacy of current DAA regimens.

Abbreviations
- DAA
direct acting antiviral
- EC50
half‐maximal effective concentration
- HCV
hepatitis C virus
- IFN
interferon
- PCR
polymerase chain reaction
- PrOD
paritaprevir/ritonavir/ombitasvir + dasabuvir
- RAS
resistance‐associated substitution
Hepatitis C virus (HCV) is a leading cause of cirrhosis and hepatocellular carcinoma, with over 70 million people chronically infected.( 1 ) The advent of potent and well‐tolerated oral direct acting antivirals (DAAs) has revolutionized treatment, providing cure rates of over 95% following 8‐24 weeks of therapy, with minimal adverse effects.( 2 ) DAAs are grouped into the NS3/4A protease inhibitors, NS5A inhibitors, and NS5B polymerase inhibitors. Most modern HCV treatment regimens combine an NS5A inhibitor with one or two drugs from other classes.
Global elimination of HCV is now achievable, and the World Health Organization has set targets of an 80% reduction in HCV incidence and a 65% reduction in HCV‐related deaths by 2030.( 3 ) One potential barrier to elimination is the emergence of antiviral resistance (particularly among people who fail therapy) with transmission of drug‐resistant viruses. Resistance‐associated substitutions (RASs) occur in all regions of the virus genome targeted by current DAAs. Although the prevalence of RASs is relatively low among persons who are treatment naive,( 4 ) the majority who fail DAA therapy develop resistant viruses in clinical trials.( 5 , 6 ) NS5A RASs are the most clinically relevant as they have been shown to reduce cure rates, particularly among difficult to treat patients, such as those with genotype 3 infection and/or cirrhosis.( 7 , 8 ) Importantly, as NS5A RASs have minimal impact on viral fitness, they may persist for years, perhaps forever.( 8 , 9 )
There is remarkably little real‐world data on HCV resistance among patients failing modern oral DAA regimens. Such data are crucial to understanding the clinical importance of HCV RASs for disease elimination.( 10 ) In March 2016, Australia became the first country to offer universal access of DAAs to all persons infected with HCV, with no restrictions for fibrosis stage or high‐risk behaviors. The Australian Government negotiated a unique volume‐based agreement for unlimited access to HCV drugs,( 11 ) the so‐called “Netflix Model.”( 12 ) Prescribing by community health care providers was encouraged to facilitate treatment scale‐up and provide access for marginalized populations. These open access models are essential to achieve HCV elimination but at the same time increase the likelihood of poor adherence among patient groups who are usually excluded from clinical trials, including those from drug and alcohol and opioid substitution therapy settings.
The introduction of the open access model in Australia led to a massive increase in community‐based DAA prescribing, with approximately 33% of persons with chronic HCV infection receiving treatment by the end of 2018.( 13 ) This makes Australia an ideal location to study the impact of DAA treatment on the emergence of resistance under an open access model. Furthermore, Australia has a higher prevalence of genotype 3 infection (~33%) than most developed countries, broadening the global relevance of our findings.
We found a high prevalence of NS5A resistance among Australian patients failing DAA therapy, particularly among those with genotype 3 infection. Almost all had failed a modern interferon (IFN)‐free DAA regimen containing an NS5A inhibitor. Multiclass drug resistance was common among those failing regimens containing both NS3 and NS5A inhibitors.
Materials and Methods
RNA Extraction and Reverse Transcription
Plasma from patients across Australia with detectable HCV RNA in plasma more than 12 weeks after completing DAA therapy was collected, transported, and stored at −80°C at Westmead Hospital, Sydney. Viral RNA extraction was performed using a QiaAMP Viral RNA kit (QIAGEN). Reverse transcription of viral RNA was performed with Superscript II (Thermo Fisher Scientific) to produce complementary DNA according to the manufacturer's instructions.
Polymerase Chain Reaction Amplification of NS3, NS5A, and NS5B
A two‐step polymerase chain reaction (PCR) was performed to amplify NS3, NS5A, and NS5B sequences using primers and protocols derived from our recent systematic review.( 14 ) The NS3 and NS5A regions were amplified and sequenced using published protocols.( 15 , 16 ) The NS5B region was amplified and sequenced using in‐house primers (forward ACCCGCTGYTTYGACTCVAC; reverse GACASGCTGWGATADATGTC). We validated the performance of our PCR assays using Australian Reference Laboratory standards and confirmed NS5A PCR sensitivity of 100% for HCV genotype 1a (10,000 IU/mL), genotype 1b (10,000 IU/mL), and genotype 3a (100,000 IU/mL). Similar sensitivities close to 100% were confirmed for the NS3 and NS5B assays.
Sequencing and Analysis
Samples were separated on a 1.5% agarose gel, stained with GelRed, and visualized under ultraviolet light. Bands corresponding to the predicted sizes of NS3, NS5A, and NS5B were excised, purified (Wizard SV Gel and PCR Clean‐Up System; Promega Corporation), and submitted for Sanger sequencing to the Australian Genome Research Facility. Sequences were analyzed for the presence of NS5A RASs using ReCALL (http://pssm.cfenet.ubc.ca/)( 17 ) and NS3 or NS5B RASs using Geno2Pheno[hcv].( 18 ) Clinically significant RASs were identified based on consensus reports and international guidelines.( 7 , 8 , 19 )
Data Analysis
Data on patient demographics, treatment history, and liver fibrosis stage were collected from request forms sent by referring clinicians. Data were analyzed using R and GraphPad Prism v8. Univariate analyses were performed using Fisher's exact test or Wilcoxon rank‐sum test as appropriate, with results presented as numbers (%) or mean, respectively; P < 0.05 was considered statistically significant.
Ethics Approval
The protocol was approved by the Western Sydney Local Health District Human Research and Ethics Committee (LNR/17/WMEAD/484). Consent was waived due to the low‐risk nature of the project and the large number of sites across the country, many with only 1 or 2 patients. This institutional ethics committee complies with the Declaration of Helsinki.
Results
Patient Cohort and Characteristics
Between January 1, 2017 and June 30, 2019, we analyzed blood samples referred from over 90 centers (representing all states and territories in Australia) from 572 patients who had failed DAA treatment. Based on details of the referring clinician, approximately 75% of samples were referred from hospitals, with the other 25% coming from community providers, sexual health clinics, or prisons.
Based on Australian prescription data, approximately 70,000 people were treated during the time frame of our study,( 20 ) with a sustained virologic response rate of approximately 96%.( 21 ) Assuming a 4% failure rate, this equates to approximately 2,800 DAA failures, so our cohort of 572 represents approximately 20% of all DAA failures in Australia, a highly representative sample.
Of the patients, 455 were men and 117 were women, with the mean age of men and women being 54.7 and 53.6 years, respectively. Rates of cirrhosis were similar in male patients (41.9%) and female patients (42.6%) (based on transient elastography or liver biopsy). The mean age of male patients with cirrhosis was 57.1 years compared with 50.7 years for those without cirrhosis (P < 0.01) (Table 1). For female patients, the average ages for those with and without cirrhosis were 59.9 years and 51.5 years, respectively (P < 0.01).
Table 1.
Summary of Patients Failing DAA Therapy, Including Age and Cirrhosis Status (Where Available)
| Sex | Total | Mean Age (years) | Cirrhosis (%) | Mean Age Cirrhosis (years) | Mean Age Without Cirrhosis (years) |
|---|---|---|---|---|---|
| Male | 455 | 53.6 | 147/351 (41.9) | 57.1 | 50.7 |
| Female | 117 | 54.7 | 40/94 (42.6) | 59.9 | 51.5 |
HCV Genotypes
The distribution of HCV genotypes among patients failing DAA therapy is summarized in Fig. 1. Of the 572 patients, 153 (26.7%) were infected with HCV genotype 1a, 34 (5.9%) with genotype 1b, 24 (4.2%) with genotype 2, 343 (60%) with genotype 3, 7 (1.2%) with genotype 4, and 11 (1.9%) with genotype 6.
Fig. 1.
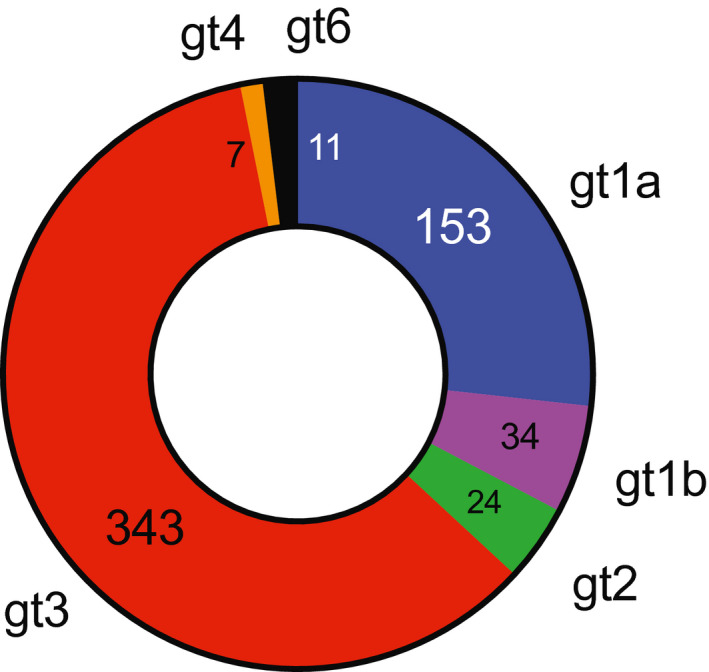
Genotype distribution of HCV isolates from patients failing DAA therapy (absolute number for each genotype). Abbreviation: gt, genotype.
DAA Regimen by HCV Genotype
For patients who failed DAAs, the regimens used for each genotype are shown in Table 2. Sofosbuvir/ledipasvir was the most common treatment regimen for genotype 1a, while sofosbuvir + daclatasvir was the most common regimen for genotype 3a. For genotype 2, most patients were treated with sofosbuvir and ribavirin as subsidized daclatasvir was not approved for genotype 2.
Table 2.
Summary of Previous DAA Treatments by HCV Genotype
| Genotype | 1a | 1b | 2 | 3 | 4 | 6 | Total |
|---|---|---|---|---|---|---|---|
| Sofosbuvir + daclatasvir | 15 | 3 | 1 | 257 | 0 | 1 | 277 |
| Sofosbuvir/ledipasvir | 94 | 16 | 0 | 22 | 1 | 3 | 136 |
| Sofosbuvir/velpatasvir | 7 | 1 | 3 | 38 | 0 | 3 | 52 |
| Elbasvir/grazoprevir | 27 | 1 | 0 | 6 | 4 | 0 | 38 |
| Sofosbuvir + ribavirin | 0 | 0 | 20 | 0 | 0 | 0 | 20 |
| PrOD | 7 | 2 | 0 | 2 | 1 | 2 | 14 |
| Glecaprevir/pibrentasvir | 1 | 1 | 0 | 4 | 0 | 0 | 6 |
| Sofosbuvir/velpatasvir/voxilaprevir | 0 | 0 | 0 | 2 | 0 | 0 | 2 |
| Other | 2 | 8 | 0 | 3 | 1 | 1 | 15 |
| Unknown | 0 | 2 | 0 | 9 | 0 | 1 | 12 |
| Total | 153 | 34 | 24 | 343 | 7 | 11 | 572 |
RAS Prevalence by HCV Genotype
Successful sequencing and RAS analysis was achieved in 556/572 (97.0%), 499/572 (87.2%), and 521/572 (91.1%) patients for NS5A, NS3, and NS5B regions, respectively. The majority of patients displayed one or more RASs in the NS5A region of the HCV genome, but the prevalence of RASs in NS3 or NS5B was much lower (Fig. 2). When calculating RAS prevalence, as the denominator we used the number of patients for whom successful sequencing was available for that gene region.
Fig. 2.
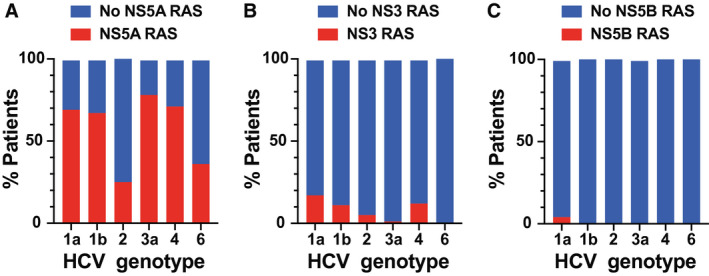
Proportion of isolates from each genotype with detectable RASs in (A) NS5A, (B) NS3, or (C) NS5B.
For NS5A (Fig. 2A), the prevalence of RASs was 72.5% (108/149) for genotype 1a, 68.8% (22/32) for genotype 1b, 26.1% (6/23) for genotype 2, 79.8% (268/336) for genotype 3, 71% (5/7) for genotype 4, and 36.4% (4/11) for genotype 6. Of note, although most patients failed a DAA combination that included an NS5A inhibitor, 19/23 patients with genotype 2 did not because they had been treated with sofosbuvir (NS5B inhibitor) and ribavirin. Among patients with genotype 2, NS5A RASs were detected in 2/4 (50%) patients who had been exposed to an NS5A inhibitor and 4/19 (21%) in those who had not.
For NS3 (Fig. 2B), the prevalence of RASs was 21.6% (30/139) for genotype 1a, 12.5% (4/32) for genotype 1b, 5.6% (1/18) for genotype 2, 1.4% (4/287) for genotype 3, 14.3% (1/7) for genotype 4, and 0% (0/8) for genotype 6. For NS5B (Fig. 2C), the prevalence of RASs was 4.2% (6/142) for genotype 1a and 0.6% (2/309) for genotype 3. NS5B RASs were not detected in other genotype infections.
NS5A RASs
RASs in the NS5A region are the most clinically relevant as all modern DAA regimens contain an NS5A inhibitor. NS5A RASs impair response to retreatment with most first‐line DAA regimens.( 22 ) Furthermore, they have minimal effect on viral fitness and may persist for months to years after DAA failure, providing potential for transmission of drug‐resistant viruses.( 8 , 23 , 24 )
A full list of NS5A RASs detected among patients with all genotypes is provided in Supporting Table S1. For patients with common genotypes (genotypes 1 or 3), we performed a more detailed analysis of NS5A RASs, summarized in Fig. 3. Among patients with HCV genotype 1a who failed DAA therapy, 108/149 (72.5%) had RASs detected at residues K24, M28, Q30, L31, H58, or Y93. Sixty‐two patients (42%) with genotype 1a infection had an RAS at a single residue, 37 patients (25%) had RASs at two different residues, 5 patients (3.3%) had RASs at three residues, and 4 (2.7%) had RASs at four residues (Fig. 3A). Among patients with HCV genotype 1b who failed DAA therapy, 22/32 (68.8%) had RASs detected at residues R30, L31, P58, or Y93. Ten patients with genotype 1b infection (31%) had an RAS at a single residue, 10 patients (31%) had RASs at two different residues, and 2 patients (6.3%) had RASs at three residues (Fig. 3B).
Fig. 3.
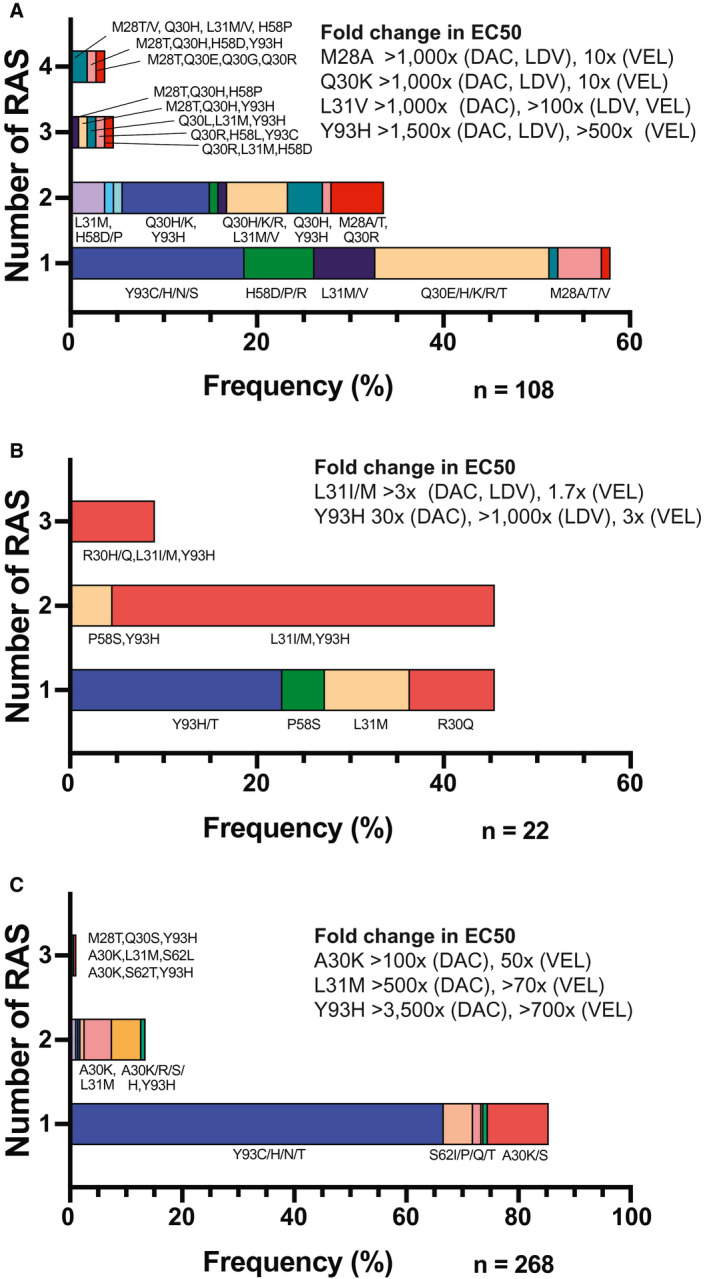
Distribution of RASs in patients infected with HCV with (A) genotype 1a, (B) genotype 1b, or (C) genotype 3, who failed DAA therapy, including sites of substitutions. Expected EC50 fold changes for common RASs are included for the most commonly used NS5A inhibitors. Abbreviations: DAC, daclatasvir; LDV, ledipasvir; VEL, velpatasvir.
Of the 336 patients with HCV genotype 3 who failed DAA therapy and had NS5A sequence data available, 327 had received treatment with an NS5A inhibitor. Overall, 268/336 patients (79.8%) had detectable RASs. Of these, 229 (68%) had an RAS on a single residue, 36 (11%) had RASs on two separate residues, and 3 (0.9%) had RASs on three separate residues (Fig. 3C). The prevalence of NS5A RASs among patients with genotype 3 who failed sofosbuvir + daclatasvir was 88% (226/257), compared to 55% (21/38) among patients who failed sofosbuvir/velpatasvir. This difference was statistically significant, with an odds ratio of 6 (P < 0.001). All 6 patients with genotype 3 who failed elbasvir/grazoprevir had NS5A RASs, as did the 2 patients who failed paritaprevir/ritonavir/ombitasvir + dasabuvir (PrOD). Two of the 4 patients with genotype 3 who failed pibrentasvir/glecaprevir (50%) had NS5A RASs; 1 had A30K and Y93H, and the other had Y93H in NS5A plus two RASs in NS3 (Y56H, D168R).
Concerning the distribution of RASs, patients with genotype 1 had a wide range of RASs across the NS5A region, while patients with genotype 3 predominantly had substitutions at residue 30 or 93 (Fig. 3C). For genotype 3, 178/336 patients (53.0%) had a single substitution at residue 93, most of these (175/178) were the Y93H substitution. Patients with genotype 3 were significantly more likely to have a single RAS than those with genotype 1 (P < 0.0001). For the small number of patients with genotype 1b, there was a trend toward multiple RASs compared to genotype 1a, but this was not statistically significant (P = 0.35).
NS3 RASs
We identified NS3 RASs in 7.8% (39/499) of patients for whom NS3 sequence data were available, mostly in patients with genotype 1 infection (Fig. 2B). Of the 39 patients with NS3 RASs, 22 (56%) had failed an oral regimen containing an NS3 inhibitor: 7 had failed PrOD, and 15 had failed elbasvir/grazoprevir, including 1 patient who failed sequential treatment with PrOD then elbasvir/grazoprevir. One patient with genotype 3a had failed glecaprevir/pibrentasvir. The range of NS3 RASs for each treatment group is summarized in Table 3.
Table 3.
List of NS3 RASs Among People Failing DAAs, Stratified by Exposure to NS3 Inhibitors
| NS3 Inhibitor Naive | PrOD | Elbasvir/ Grazoprevir | |||
|---|---|---|---|---|---|
| RAS | Frequency | RAS | Frequency | RAS | Frequency |
| L30S | 1 | Q80K | 1 | V55A | 1 |
| V55A | 1 | R155K | 1 | Y56F | 1 |
| Y56F | 1 | V55A, Q80K | 1 | Y56H | 2 |
| Q80K | 4 | V55A, D168A | 1 | Q80K | 1 |
| Q80L | 1 | Q80K, R155T, D168A | 1 | D168A | 2 |
| S122G | 4 | V36M, Q80K, D168A | 1 | D168L | 1 |
| D168K | 1 | Y56H, S122N, D168V | 1 | I170V | 1 |
| Q168R | 1 | A156T, D168N | 1 | ||
| I170V | 1 | V55A, D168A | 1 | ||
| T54S, A155K | 1 | Y56H, D168A | 1 | ||
| D168H, V170A | 1 | Y56H, D168V | 1 | ||
| V36M, Y56H, D168A | 1 | ||||
| V55A, Q80K, D168A | 1 | ||||
| Total | 17 | Total | 7 | Total | 15 |
| RAS Prevalence | 17/432 (3.9%) | 7/14 (50%) | 15/43 (35%) | ||
There was an increased rate of NS3 resistance among patients exposed to an oral regimen containing an NS3 inhibitor, with an odds ratio of 13.4 (P < 0.0001). Overall, the prevalence of NS3 RASs was 35% (22/62) among patients who had been exposed to an oral regimen containing an NS3 inhibitor compared with 3.9% (17/437) for patients who had not. For patients with genotype 1a infection, among 33 patients treated with an oral regimen containing an NS3 inhibitor, the prevalence of NS3 RASs was 58% (19/33) compared to 10% (11/106) for those who had not, an odds ratio of 11.7 (P < 0.0001). Comparing regimens, the prevalence of NS3 RASs was 50% (7/14) for patients failing PrOD, 35% (15/43) for patients failing elbasvir/grazoprevir, and 17% (1/6) for patients failing glecaprevir/pibrentasvir.
NS5B RASs
Among the 521 patients for whom NS5B data are available, NS5B RASs were detected in 8 patients (1.5%); 6 were in patients with genotype 1a (4% prevalence) and 2 with genotype 3 (1%) (Fig. 2C).
Among the 14 patients exposed to the non‐nucleotide NS5B inhibitor dasabuvir (in PrOD), 3 (21%) had detectable dasabuvir RASs in NS5B. The first patient had Y561I and S556G substitutions, the second had a single RAS (S556G), while the third patient had N444D and A553X RASs.
There was a single sofosbuvir RAS detected in a patient with genotype 3a who had failed treatment with sofosbuvir and daclatasvir. The clinically significant sofosbuvir RAS S282T was detected 15 weeks after completing treatment but was no longer detectable by Sanger sequencing 12 weeks later. A recent report suggested the common A150V polymorphism and other variants may confer sofosbuvir resistance for genotype 3.( 25 ) We did not see a significantly increased frequency of A150V or other listed variants compared to untreated patients in the international Surveillance of Hepatitis‐C Antiviral Resistance, Epidemiology and methoDologies (SHARED) database (data not shown), which includes patients from our cohort.
Multiclass Resistance
Of patients who failed all‐oral DAA regimens that contained both an NS3 and NS5A inhibitor, 32% (20/62) had both NS3 and NS5A RASs detected compared with 3.1% (16/510) among those not exposed to both an NS3 and NS5A inhibitor. This demonstrates a dramatically increased risk of multiclass resistance among patients exposed to both NS3 and NS5A inhibitors, with an odds ratio of 14.7 (P < 0.001).
Looking at specific regimens, of the patients failing elbasvir/grazoprevir who had NS3 RASs, 14/15 (93%) also had NS5A RASs. Of the patients who failed PrOD, 7/14 (50%) had NS3 RASs. All these patients had genotype 1a infection, and all but 1 had multiclass resistance. Three patients had multiple RASs in the NS3 and NS5A regions, 1 patient had NS3 and NS5B RASs, and 2 patients had RASs in all three regions targeted by DAAs (NS3, NS5A and NS5B) (Table 4).
Table 4.
Multiclass Resistance Among Patients With HCV Genotype 1a Who Failed Treatment With PrOD and Ribavirin
| Treatment 1 | Treatment 2 | Genotype | NS3 RAS | NS5A RAS | NS5B RAS |
|---|---|---|---|---|---|
| PrOD + ribavirin | Sofosbuvir/ledipasvir | 1a | Q80K | A553D | |
| PrOD + ribavirin | 1a | Q80K, R155T, D168A | M28T, Q30H, H58D, Y93H | S556G | |
| PrOD + ribavirin | 1a | R155K | |||
| IFN, ribavirin, telaprevir | PrOD + ribavirin | 1a | V36M, Q80K, D168A | Q30R | Y561I, S556G |
| PrOD + ribavirin | Elbasvir/grazoprevir | 1a | V55A, D168A | Q30R, L31V | |
| PrOD + ribavirin | 1a | V55A, Q80K | H58P | ||
| PrOD + ribavirin | 1a | Y56H, S122N, D168V | Q30H, Y93H |
Among patients with genotype 3 who failed glecaprevir/pibrentasvir, 1 of the 4 had dual‐class resistance with the Y93H RAS in NS5A and two RASs in NS3 (Y56H, D168R). The single patient with genotype 3 infection in whom the sofosbuvir‐specific RAS (S282T) in NS5B was detected also had high level NS5A resistance with the Y93H RAS.
Effect of Cirrhosis on Antiviral Resistance Development
To look for a possible effect of cirrhosis on the development of HCV antiviral resistance, we compared the genotype distribution and number of RASs between patients with or without cirrhosis who had failed DAA therapy. There was no apparent interaction between HCV genotype and cirrhosis for DAA failure. The relative genotype distribution did not differ between patients with or without cirrhosis, for either male (Fig. 4A) or female (Fig. 4B) patients. There was no apparent difference in the number of RASs between patients with and without cirrhosis for either male (Fig. 4C) or female (Fig. 4D) patients.
Fig. 4.
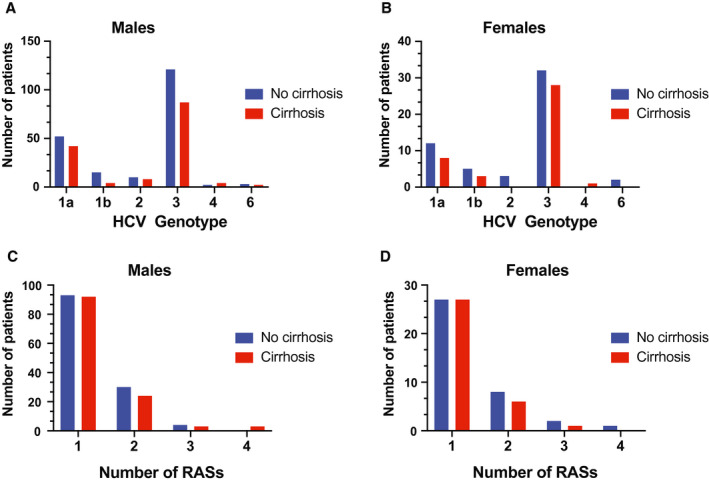
Distribution of HCV genotypes among (A) male patients and (B) female patients failing DAA therapy, stratified by the presence or absence of cirrhosis. Number of RASs in virus isolated from (C) male patients and (D) female patients after failing DAA therapy, stratified by the presence or absence of cirrhosis.
Discussion
To achieve global hepatitis C elimination, increased diagnosis, open access to treatment, and streamlined treatment pathways are required.( 26 ) High cure rates in the real world have been confirmed for IFN‐free DAA regimens,( 27 , 28 ) but elimination targets could be compromised by the emergence and transmission of drug‐resistant virus, so surveillance and monitoring of real‐world antiviral resistance trends are crucial.( 10 , 29 ) In this, the largest real‐world study of patients failing a currently recommended IFN‐free regimen, 525/572 (92%) patients received an NS5A inhibitor, including 58 with one of the new pangenotype regimens, velpatasvir/sofosbuvir or pibrentasvir/glecaprevir. Sixty percent of patients (343/572) had genotype 3 infection, the largest cohort to date of this difficult‐to‐cure genotype. Importantly, our data represent approximately 20% of all patients in Australia failing DAAs, a highly representative sample.
A recent real‐world study by Dietz and colleagues( 30 ) included 626 European patients who had failed DAA therapy (73 with genotype 3), but only 376 received an NS5A inhibitor, the rest being treated with sofosbuvir plus either simeprevir (NS3 inhibitor), ribavirin, and/or pegylated IFN. Of note, no patient in that study received either of the new pangenotype regimens.( 30 ) The next largest study to date included 197 DAA failures, but only 15 received a DAA regimen containing an NS5A inhibitor.( 31 )
We detected NS5A RASs in approximately 72% and 80% of patients who failed therapy with genotype 1 and 3 infections, respectively. Surprisingly, RAS prevalence was lower than in the registration trials. In phase 2/3 trials of ledipasvir/sofosbuvir for HCV genotype 1, the prevalence of RASs among patients who completed 12 to 24 weeks of therapy was 95% to 100%,( 5 ) and in the phase 3 trial of sofosbuvir plus daclatasvir for genotype 3, all patients who relapsed had NS5A RASs.( 6 ) One explanation for the lower RAS prevalence in real‐world studies is reduced treatment adherence and/or completion because RAS prevalence depends on duration of therapy. In the ledipasvir/sofosbuvir trials, for patients who failed 6, 8, 12, or 24 weeks therapy, the prevalence of NS5A RASs was 37.5%, 66.7%, 94.7%, or 100%, respectively.( 5 ) Thus, relapse may be due to inadequate treatment after short courses, while for longer courses, failure was likely due to antiviral resistance. Another explanation for lower RAS prevalence in real‐world studies is that some patients are likely to have been reinfected with wild‐type HCV rather than relapsing with resistant viruses.
This is the first, large, real‐world study examining the prevalence of RASs among patients failing the pangenotype regimen sofosbuvir/velpatasvir. We observed a lower prevalence of NS5A RASs among patients with genotype 3 who failed sofosbuvir/velpatasvir (55%) than sofosbuvir + daclatasvir (88%). This contrasts with results from the sofosbuvir/velpatasvir registration trials in which all patients with genotype 3 (12/12) who failed therapy had NS5A RASs.( 32 ) Our results may reflect reduced treatment adherence and/or reinfection in the community for patients taking sofosbuvir/velpatasvir due to its likely higher use among high‐risk patients. Between March 2016 and August 2017, there was a dramatic change in HCV prescribing in Australia, with increased community prescribing; the proportion of DAAs prescribed by specialists fell from 80% to 33%, with a corresponding increase in family practice prescribing, from 8% to 41%.( 20 ) In the first 6 months, many patients prescribed DAAs were already engaged with specialist liver clinics and had been waiting for DAA access.( 33 ) By the time sofosbuvir/velpatasvir became available, more patients were being prescribed DAAs in the community, with potentially increased risk of poor treatment adherence, premature cessation, and/or reinfection. Previous community treatment studies in Australia confirmed that adherence was lower for patients treated later after DAAs became available compared to “early adopters” who may have been more motivated or adherent to treatment.( 34 ) Consistent with this, in our cohort there was a trend toward higher resistance rates in patients referred by community prescribers compared to hospital prescribers, but this did not reach statistical significance. This highlights the importance of monitoring treatment adherence and reinfection rates among high‐risk groups, particularly during treatment scale‐up with open access models.
A prominent finding was the divergence between genotypes 1a and 3a in the NS5A RAS distribution. Patients with genotype 1a failing DAA therapy were more likely to have multiple NS5A RASs, particularly at positions 28, 30, 31, 58, and 93. The relatively high prevalence (12%) of NS5A RASs at position 58 may be clinically significant as H58D confers high‐level resistance to most NS5A inhibitors, including velpatasvir, with possible resistance against pibrentasvir.( 35 ) This difference may reflect geographic variations in baseline RAS prevalence as we have previously observed a 4% prevalence of H58 RAS among untreated Australian patients with genotype 1a( 36 ) compared to 0%‐2% in global and European patients.( 16 , 37 ) In contrast to the wide range of RASs for genotype 1a, patients with genotype 3a were more likely to have a single NS5A RAS, with Y93H found in 72%. This is similar to the 66% prevalence in the European study( 30 ) and may reflect a relatively high baseline prevalence of Y93H (9%) among patients with genotype 3a who are treatment naive.( 37 ) The very high prevalence of Y93H among treatment failures is clinically important as Y93H confers high levels of resistance against most NS5A inhibitors, with a 3,500‐fold reduction in half‐maximal effective concentration (EC50) for daclatasvir and over 700‐fold reduction in EC50 for velpatasvir.( 38 )
For patients with genotype 2 infection, the low proportion of NS5A RASs (26%) is not surprising as only 4/24 (16%) were treated with an NS5A inhibitor, the rest receiving sofosbuvir + ribavirin, providing no selective pressure for NS5A RASs. Among patients with genotype 4, NS5A RASs were detected in 71% (5/7), 2 of whom failed elbasvir/grazoprevir, 1 ledipasvir/sofosbuvir, 1 PrOD, and 1 sofosbuvir + ribavirin. The 2 patients without NS5A RASs failed elbasvir/grazoprevir. One patient with genotype 4r who failed elbasvir/grazoprevir had dual‐class resistance, with three NS5A RASs (L28M, L30R, and Y93H) and two NS3 RASs (A156T, D168N). This is similar to the European and Italian studies, where high rates of NS5A resistance were seen among patients with genotype 4 failing DAAs, with dual‐class resistance in patients failing PrOD, but no patients in those studies received elbasvir/grazoprevir.( 30 , 31 ) For genotype 6, the low prevalence of NS5A RASs (30%) may be due to patients failing suboptimal DAA regimens prescribed following incorrect genotype identification. Laboratory identification of genotype 6 can be difficult, and on early commercial genotype assays, genotype 6 was frequently misidentified as genotype 1b.( 39 ) This highlights the importance of considering genotype 6 in patients from endemic countries of South‐East Asia, such as Cambodia and Thailand, to guide appropriate pangenotypic regimens.
An important finding was the high prevalence (33%) of multiclass drug resistance among patients who failed an all‐oral regimen containing both an NS3 and NS5A inhibitor. This is likely an underestimate as data were not available on the timing of samples relative to treatment completion date and RASs in the NS3 region usually become undetectable after several weeks to months due to reduced viral fitness.( 40 ) Based on national prescription data, of the ~70,000 patients in Australia treated with IFN‐free DAAs between March 2016 and December 2018, approximately 7,000 (10%) received a regimen containing both an NS3 and NS5A inhibitor (PrOD, 1,000; elbasvir/grazoprevir, 4,000; grazoprevir/pibrentasvir, 2,000).( 20 ) Assuming a 96% cure rate based on real‐world Australian data,( 21 ) approximately 280 patients (4% of 7,000) would have failed an NS3/NS5A inhibitor regimen, meaning around 93 (33% of 280) are likely to have dual‐class resistance.
The clinical impact of multiclass drug resistance on retreatment outcomes is unclear, but concerning data are starting to emerge. In phase 3 trials, retreatment with glecaprevir (NS3 inhibitor) and pibrentasvir (NS5A inhibitor) cured >90% of patients who failed a DAA regimen containing either an NS5A or NS3 inhibitor.( 41 ) However, cure rates were lower (~80% overall) for patients previously exposed to both NS3 and NS5A inhibitors and fell to 25% for patients with detectable RASs in both NS3 and NS5A.( 41 ) The triple combination of sofosbuvir/velpatasvir/voxilaprevir is highly effective for retreating patients who fail DAA therapy, with cure rates >95% in phase 3 studies,( 42 ) but real‐world studies suggest reduced cure rates in people with genotype 3 infection, cirrhosis, and those who had failed sofosbuvir/velpatasvir.( 43 , 44 ) In the phase 3 trials, there was no apparent impact of RASs, but only a minority of patients had multiclass resistance and 1 patient with virologic breakthrough on treatment had multiple RASs in NS5A (Q30T, L31M, Y93H) and NS3 (Q80K).( 42 ) Thus, further real‐world studies are warranted.
A single patient with genotype 3 had a high level sofosbuvir RAS (S282T). This was detected 15 weeks after completing sofosbuvir + daclatasvir but not 12 weeks later, consistent with the known impact of S282T on viral fitness.( 8 ) Of concern, compensatory mutations can restore viral fitness in the context of S282T, providing potential for transmission of sofosbuvir resistance in high‐risk settings. This has been demonstrated in vitro for genotypes 3 and 6,( 45 , 46 ) including a replication‐competent virus that was resistant to all three classes of pangenotype DAAs, pibrentasvir (NS3), velpatasvir (NS5A), and sofosbuvir (NS5B).( 45 ) Chronic infection with sofosbuvir‐resistant virus (S282T) has now been confirmed in a high‐risk patient with genotype 4d.( 47 )
Global elimination of hepatitis C requires widespread treatment scale‐up and open access to DAAs, strategies that also increase the risk of emergence and transmission of drug‐resistant viruses. The present study confirms a high prevalence of NS5A resistance among people who fail IFN‐free DAA therapy and high rates of multiclass drug resistance in those exposed to both NS3 and NS5A inhibitors. When retreating patients in the community, it can be difficult to obtain an accurate history of prior DAA exposure, so RAS testing may be helpful to guide selection of an appropriate salvage regimen. Another pragmatic approach to reducing multiclass resistance would be to restrict first‐line treatment to regimens containing only NS5A and NS5B inhibitors, reserving NS3 inhibitors for salvage therapy.
Supporting information
TableS1
Supported by the National Health and Medical Research Council of Australia (grant 1053206 to AL, GD, JG, MD, ET, RB, and TA and a postgraduate scholarship to A.O.), Australian Centre for HIV and Hepatitis Virology Research, University of Sydney (grant to MD, RB, TA), Western Sydney Local Health District Research Education Network (grant to MD, ET), and the Robert W. Storr bequest to the Sydney Medical Foundation (University of Sydney), (Sydney Medical School Accelerator grant to MD).
Potential conflict of interest: Dr. Douglas advises, is on the speakers' bureau for Gilead, AbbVie, and Merck, and has received grants from Gilead and AbbVie. Dr. George advises, is on the speakers’ bureau for, and received grants from Gilead; he advises and is on the speakers’ bureau for AbbVie and MSD. Dr. Dore advises, is on the speakers’ bureau for, and received grants from Gilead, AbbVie, and Merck. Dr. Lloyd received grants from Gilead and AbbVie. The other authors have nothing to report.
References
Author names in bold designate shared co‐first authorship.
- 1. The Polaris Observatory HCV Collaborators . Global prevalence and genotype distribution of hepatitis C virus infection in 2015: a modelling study. Lancet Gastroenterol Hepatol 2017;2:161‐176. [DOI] [PubMed] [Google Scholar]
- 2. Chung RT, Baumert TF. Curing chronic hepatitis C–the arc of a medical triumph. N Engl J Med 2014;370:1576‐1568. [DOI] [PubMed] [Google Scholar]
- 3. World Health Organization . Global Health Sector Strategy on Viral Hepatitis 2016–2021: towards ending viral hepatitis. Geneva, Switzerland: World Health Organisation; 2016. [Google Scholar]
- 4. Chen ZW, Li H, Ren H, Hu P. Global prevalence of pre‐existing HCV variants resistant to direct‐acting antiviral agents (DAAs): mining the GenBank HCV genome data. Sci Rep 2016;6:20310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Wyles D, Dvory‐Sobol H, Svarovskaia ES, Doehle BP, Martin R, Afdhal NH, et al. Post‐treatment resistance analysis of hepatitis C virus from phase II and III clinical trials of ledipasvir/sofosbuvir. J Hepatol 2017;66:703‐710. [DOI] [PubMed] [Google Scholar]
- 6. Nelson DR, Cooper JN, Lalezari JP, Lawitz E, Pockros PJ, Gitlin N, et al.; ALLY‐3 Study Team . All‐oral 12‐week treatment with daclatasvir plus sofosbuvir in patients with hepatitis C virus genotype 3 infection: ALLY‐3 phase III study. Hepatology 2015;61:1127‐1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lontok E, Harrington P, Howe A, Kieffer T, Lennerstrand J, Lenz O, et al. Hepatitis C virus drug resistance‐associated substitutions: state of the art summary. Hepatology 2015;62:1623‐1632. [DOI] [PubMed] [Google Scholar]
- 8. Pawlotsky JM. Hepatitis C virus resistance to direct‐acting antiviral drugs in interferon‐free regimens. Gastroenterology 2016;151:70‐86. [DOI] [PubMed] [Google Scholar]
- 9. Wyles D, Mangia A, Cheng W, Shafran S, Schwabe C, Ouyang W, et al. Long‐term persistence of HCV NS5A resistance‐associated substitutions after treatment with the HCV NS5A inhibitor, ledipasvir, without sofosbuvir. Antivir Ther 2018;23:229‐238. [DOI] [PubMed] [Google Scholar]
- 10. Doyle A, Feld JJ. Overcoming the resistance to resistance testing: collecting the data. Liver Int 2017;37:506‐508. [DOI] [PubMed] [Google Scholar]
- 11. Alcorn K. Australia shows an alternative to rationing hepatitis C treatment. NAM AIDSMAP; 2016. https://www.aidsmap.com/news/jun-2016/australia-shows-alternative-rationing-hepatitis-c-treatment. [Google Scholar]
- 12. Moon S, Erickson E. Universal medicine access through lump‐sum remuneration‐Australia's approach to hepatitis C. N Engl J Med 2019;380:607‐610. [DOI] [PubMed] [Google Scholar]
- 13. The Kirby Institute . Monitoring Hepatitis C Treatment Uptake in Australia (Issue 9). University of New South Wales, The Kirby Institute; 2018. https://kirby.unsw.edu.au/report/monitoring-hepatitis-c-treatment-uptake-australia-issue-9-july-2018. [Google Scholar]
- 14. Bartlett SR, Grebely J, Eltahla AA, Reeves JD, Howe AYM, Miller V, et al. Sequencing of hepatitis C virus for detection of resistance to direct‐acting antiviral therapy: a systematic review. Hepatol Commun 2017;1:379‐390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Besse B, Coste‐Burel M, Bourgeois N, Feray C, Imbert‐Marcille BM, Andre‐Garnier E. Genotyping and resistance profile of hepatitis C (HCV) genotypes 1–6 by sequencing the NS3 protease region using a single optimized sensitive method. J Virol Methods 2012;185:94‐100. [DOI] [PubMed] [Google Scholar]
- 16. Lindstrom I, Kjellin M, Palanisamy N, Bondeson K, Wesslen L, Lannergard A, et al. Prevalence of polymorphisms with significant resistance to NS5A inhibitors in treatment‐naive patients with hepatitis C virus genotypes 1a and 3a in Sweden. Infect Dis (Lond) 2015;47:555‐562. [DOI] [PubMed] [Google Scholar]
- 17. Rhee SY, Blanco JL, Jordan MR, Taylor J, Lemey P, Varghese V, et al. Geographic and temporal trends in the molecular epidemiology and genetic mechanisms of transmitted HIV‐1 drug resistance: an individual‐patient‐ and sequence‐level meta‐analysis. PLoS Med 2015;12:e1001810. Erratum in: PLoS Med 2015;12:e1001845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kalaghatgi P, Sikorski AM, Knops E, Rupp D, Sierra S, Heger E, et al. Geno2pheno[HCV] ‐ a web‐based interpretation system to support hepatitis C treatment decisions in the era of direct‐acting antiviral agents. PLoS One 2016;11:e0155869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. European Association for the Study of the Liver . EASL recommendations on treatment of hepatitis C 2018. J Hepatol 2018;69:461‐511. [DOI] [PubMed] [Google Scholar]
- 20. The Kirby Institute . Monitoring Hepatitis C Treatment Uptake in Australia (Issue 10). University of New South Wales, The Kirby Institute; 2019. https://kirby.unsw.edu.au/report/monitoring-hepatitis-c-treatment-uptake-australia-issue-10-june-2019. [Google Scholar]
- 21. The Kirby Institute . Real‐World Efficacy of Antiviral Therapy in Chronic Hepatitis C (REACH‐C) in Australia (Issue 2). University of New South Wales, The Kirby Institute; 2018. https://kirby.unsw.edu.au/. [Google Scholar]
- 22. Lawitz E, Flamm S, Yang JC, Pang PS, Zhu Y, Svarovskaia E, et al. O005: retreatment of patients who failed 8 or 12 weeks of ledipasvir/sofosbuvir‐based regimens with ledipasvir/sofosbuvir for 24 weeks. J Hepatol 2015;62(Suppl. 2):S192. [Google Scholar]
- 23. Franco S, Tural C, Nevot M, Molto J, Rockstroh JK, Clotet B, et al. Detection of a sexually transmitted hepatitis C virus protease inhibitor‐resistance variant in a human immunodeficiency virus‐infected homosexual man. Gastroenterology 2014;147:599‐601.e1. [DOI] [PubMed] [Google Scholar]
- 24. Abravanel F, Metivier S, Chauveau M, Peron JM, Izopet J. Transmission of HCV NS5A inhibitor‐resistant variants among HIV‐infected men who have sex with men. Clin Infect Dis 2016;63:1271‐1272. [DOI] [PubMed] [Google Scholar]
- 25. Wing PAC, Jones M, Cheung M, DaSilva S, Bamford C, Jason Lee WY, et al. Amino acid substitutions in genotype 3a hepatitis C virus polymerase protein affect responses to sofosbuvir. Gastroenterology 2019;157:692‐704.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Thomas DL. Global elimination of chronic hepatitis. N Engl J Med 2019;380:2041‐2050. [DOI] [PubMed] [Google Scholar]
- 27. Calleja JL, Crespo J, Rincon D, Ruiz‐Antoran B, Fernandez I, Perello C, et al.; Spanish Group for the Study of the Use of Direct‐acting Drugs Hepatitis C Collaborating Group . Effectiveness, safety and clinical outcomes of direct‐acting antiviral therapy in HCV genotype 1 infection: results from a Spanish real‐world cohort. J Hepatol 2017;66:1138‐1148. [DOI] [PubMed] [Google Scholar]
- 28. Ioannou GN, Beste LA, Chang MF, Green PK, Lowy E, Tsui JI, et al. Effectiveness of sofosbuvir, ledipasvir/sofosbuvir, or paritaprevir/ritonavir/ombitasvir and dasabuvir regimens for treatment of patients with hepatitis C in the Veterans Affairs national health care system. Gastroenterology 2016;151:457‐471.e5. Erratum in: Gastroenterology 2016;151:1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Popping S, Cento V, Garcia F, Ceccherini‐Silberstein F, Seguin‐Devaux C, Vijver DA, et al. The need for a European hepatitis C programme monitoring resistance to direct‐acting antiviral agents in real life to eliminate hepatitis C. J Virus Erad 2018;4:179‐181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Dietz J, Susser S, Vermehren J, Peiffer KH, Grammatikos G, Berger A, et al.; European HCV Resistance Study Group . Patterns of resistance‐associated substitutions in patients with chronic HCV infection following treatment with direct‐acting antivirals. Gastroenterology 2018;154:976‐988.e974. [DOI] [PubMed] [Google Scholar]
- 31. Di Maio VC, Cento V, Lenci I, Aragri M, Rossi P, Barbaliscia S, et al.; HCV Italian Resistance Network Study Group . Multiclass HCV resistance to direct‐acting antiviral failure in real‐life patients advocates for tailored second‐line therapies. Liver Int 2017;37:514‐528. [DOI] [PubMed] [Google Scholar]
- 32. Hezode C, Reau N, Svarovskaia ES, Doehle BP, Shanmugam R, Dvory‐Sobol H, et al. Resistance analysis in patients with genotype 1–6 HCV infection treated with sofosbuvir/velpatasvir in the phase III studies. J Hepatol 2018;68:895‐903. [DOI] [PubMed] [Google Scholar]
- 33. Hajarizadeh B, Grebely J, Matthews GV, Martinello M, Dore GJ. Uptake of direct‐acting antiviral treatment for chronic hepatitis C in Australia. J Viral Hepat 2018;25:640‐648. [DOI] [PubMed] [Google Scholar]
- 34. Read P, Gilliver R, Kearley J, Lothian R, Cunningham EB, Chronister KJ, et al. Treatment adherence and support for people who inject drugs taking direct‐acting antiviral therapy for hepatitis C infection. J Viral Hepat 2019;26:1301‐1310. [DOI] [PubMed] [Google Scholar]
- 35. Sorbo MC, Cento V, Di Maio VC, Howe AYM, Garcia F, Perno CF, et al. Hepatitis C virus drug resistance associated substitutions and their clinical relevance: update 2018. Drug Resist Updat 2018;37:17‐39. Erratum in: Drug Resist Updat 2018. [DOI] [PubMed] [Google Scholar]
- 36. Ong AT, Tay E, Dwyer DE, George J, Douglas MW. Pre‐treatment antiviral resistance in Australians with chronic hepatitis C: prevalence of NS3 and NS5A resistance data in the state of New South Wales. Antivir Ther 2019;24:281‐290. [DOI] [PubMed] [Google Scholar]
- 37. Palanisamy N, Kalaghatgi P, Akaberi D, Lundkvist A, Chen ZW, Hu P, et al. Worldwide prevalence of baseline resistance‐associated polymorphisms and resistance mutations in HCV against current direct‐acting antivirals. Antivir Ther 2018;23:485‐493. [DOI] [PubMed] [Google Scholar]
- 38. Dvory‐Sobol H, Han B, Lu J, Yu M, Beran RK, Cheng G, et al. In vitro resistance profile of hepatitis C virus NS5A inhibitor velpatasvir in genotypes 1 to 6. J Viral Hepat 2019;26:991‐1001. [DOI] [PubMed] [Google Scholar]
- 39. Chinchai T, Labout J, Noppornpanth S, Theamboonlers A, Haagmans BL, Osterhaus AD, et al. Comparative study of different methods to genotype hepatitis C virus type 6 variants. J Virol Methods 2003;109:195‐201. [DOI] [PubMed] [Google Scholar]
- 40. Pawlotsky JM. Treatment failure and resistance with direct‐acting antiviral drugs against hepatitis C virus. Hepatology 2011;53:1742‐1751. [DOI] [PubMed] [Google Scholar]
- 41. Poordad F, Pol S, Asatryan A, Buti M, Shaw D, Hezode C, et al. Glecaprevir/pibrentasvir in patients with hepatitis C virus genotype 1 or 4 and past direct‐acting antiviral treatment failure. Hepatology 2018;67:1253‐1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Sarrazin C, Cooper CL, Manns MP, Reddy KR, Kowdley KV, Roberts SK, et al. No impact of resistance‐associated substitutions on the efficacy of sofosbuvir, velpatasvir, and voxilaprevir for 12weeks in HCV DAA‐experienced patients. J Hepatol 2018;69:1221‐1230. [DOI] [PubMed] [Google Scholar]
- 43. Belperio PS, Shahoumian TA, Loomis TP, Backus LI. Real‐world effectiveness of sofosbuvir/velpatasvir/voxilaprevir in 573 direct‐acting antiviral experienced hepatitis C patients. J Viral Hepat 2019;26:980‐990. [DOI] [PubMed] [Google Scholar]
- 44. Llaneras J, Riveiro‐Barciela M, Lens S, Diago M, Cachero A, Garcia‐Samaniego J, et al. Effectiveness and safety of sofosbuvir/velpatasvir/voxilaprevir in patients with chronic hepatitis C previously treated with DAAs. J Hepatol 2019;71:666‐672. [DOI] [PubMed] [Google Scholar]
- 45. Pham LV, Ramirez S, Gottwein JM, Fahnoe U, Li YP, Pedersen J, et al. HCV genotype 6a escape from and resistance to velpatasvir, pibrentasvir, and sofosbuvir in robust infectious cell culture models. Gastroenterology 2018;154:2194‐2208.e12. [DOI] [PubMed] [Google Scholar]
- 46. Ramirez S, Mikkelsen LS, Gottwein JM, Bukh J. Robust HCV genotype 3a infectious cell culture system permits identification of escape variants with resistance to sofosbuvir. Gastroenterology 2016;151:973‐985.e2. [DOI] [PubMed] [Google Scholar]
- 47. Newsum AM, Molenkamp R, van der Meer JT, Rebers SP, Prins M, van der Valk M, et al. Persistence of NS5B‐S282T, a sofosbuvir resistance‐associated substitution, in a HIV/HCV‐coinfected MSM with risk of onward transmission. J Hepatol 2018;69:968‐970. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
TableS1


