Figure 2.
Tumor Education Signatures of TAMs in BrM
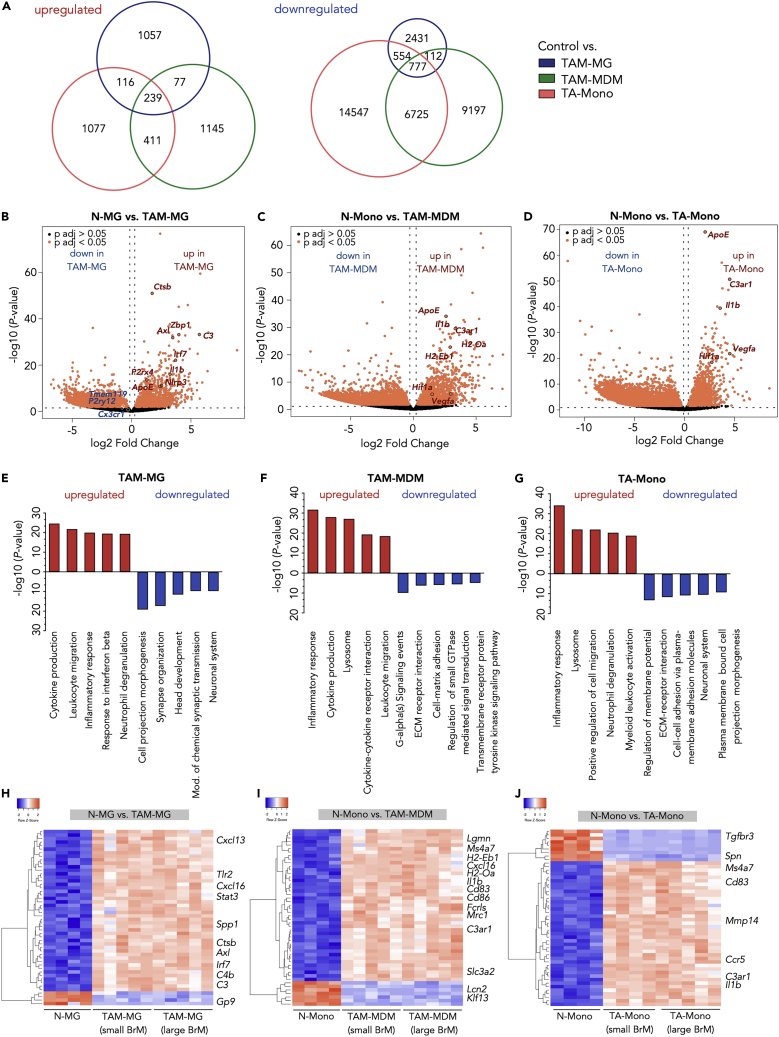
(A) Euler diagrams depict the number of unique and shared upregulated (left panel) or downregulated (right panel) DEGs in control versus TAM-MG, or TAM-MDM, or TA-Mono.
(B–D) Volcano plot of control cells versus BrM-associated cells in (B) microglia, (C) MDM, and (D) monocytes. Cutoffs: ± 1 log2 fold change and adjusted p value <0.05.
(E–G) Functional gene annotation of altered cellular pathways in (E) TAM-MG, (F) TAM-MDM, and (G) TA-Mono compared with control cell types from tumor-free animals. Cutoffs: ± 1, log2 fold change and adjusted p value <0.05. All DEGs or top 3,000 DEGs based on adjusted p values were subjected to analysis.
(H–J) Unsupervised clustering of the top 50 DEGs in control versus BrM-associated (small and large stage) TAMs for (H) microglia, (I) MDM, and (J) monocytes. n = 4–5 replicates for each condition.