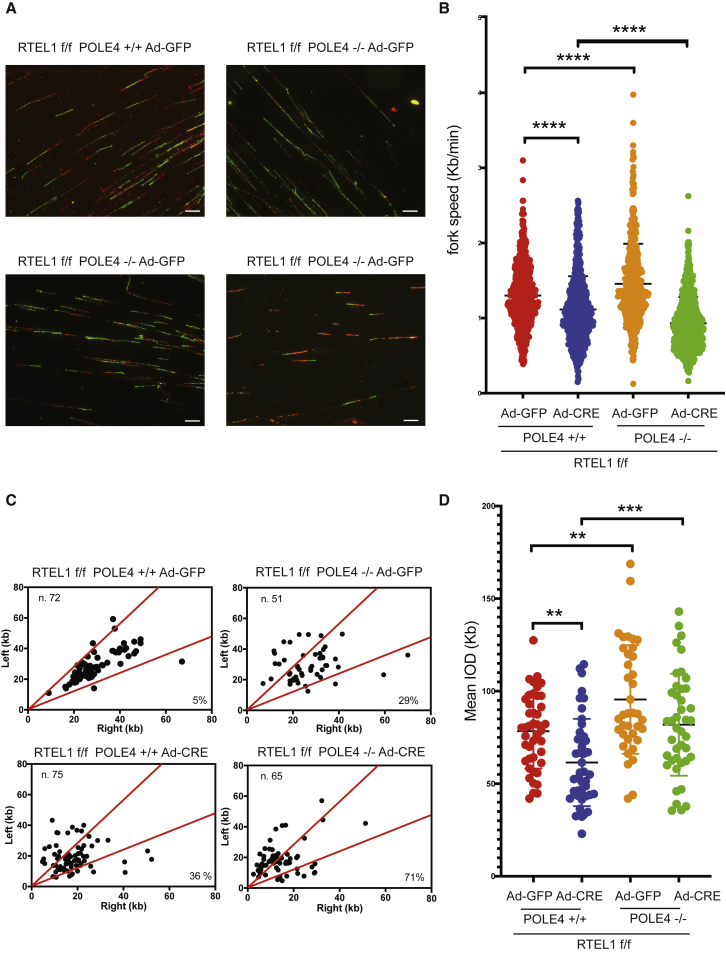
Figure 7.
Impaired Fork Progression and Increased Fork Stalling in RTEL1-POLE4 Double Knockout Cells
(A) Representative DNA fiber immunofluorescence from RTEL1F/FPOLE4+/+ and RTEL1F/FPOLE4−/− MEFs infected with GFP-CRE or empty GFP.
(B) Bar graphs showing replication fork speed (measured as IdU track length/min) from RTEL1F/FPOLE4+/+ and RTEL1F/FPOLE4−/− MEFs transduced or not with CRE. ∗∗∗∗p < 0.0001.
(C). Analysis of replication fork symmetry from RTEL1F/FPOLE4+/+ and RTEL1F/FPOLE4−/− MEFs infected with GFP-CRE or empty GFP. Total number of newly established replication forks analyzed is indicated.
(D) Bar graphs showing inter-origin distance measurements from RTEL1F/FPOLE4+/+ and RTEL1F/FPOLE4−/− MEFs transduced or not with CRE recombinase (∗∗p < 0.01, ∗∗∗p < 0.001). Error bars represent standard deviation (SD) of the mean. Scale bars, 10 μm.