Fig. 4.
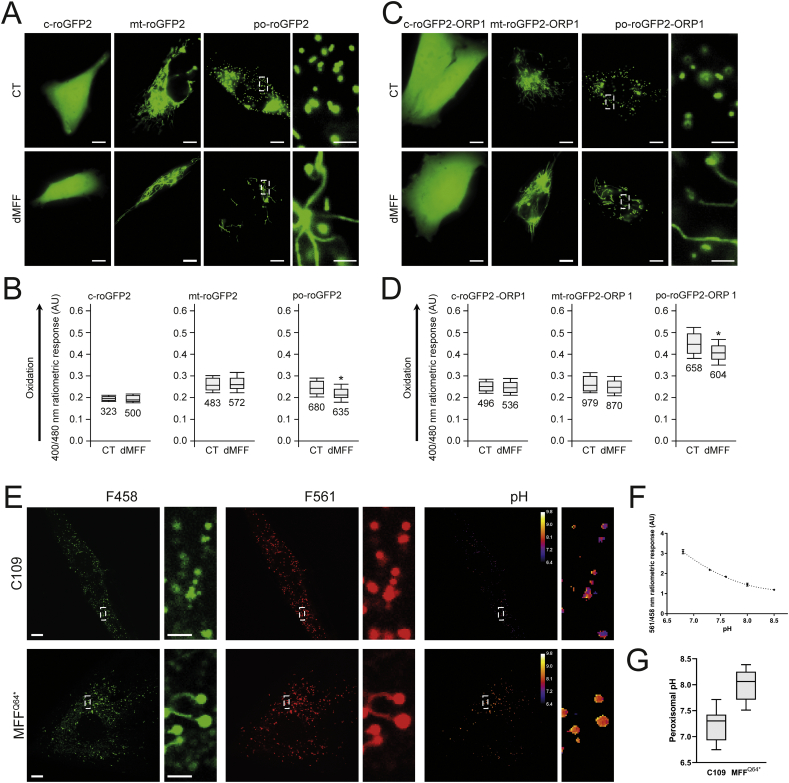
Peroxisomal redox state and pH levels are altered in MFF-deficient fibroblasts. Control (CT) or MFF-deficient (dMFF) SV40 large T antigen-transformed human fibroblasts (HUFs-T) were transfected with a plasmid encoding cytosolic (c-), mitochondrial (mt-) or peroxisomal (po-) roGFP2 (A, B) or roGFP2-ORP1 (C, D). (A) Distribution patterns of the respective roGFP2 proteins. Higher magnification view of po-roGFP2 is shown. (B) Box plot representations of the 400/480 nm fluorescence response ratios of the respective roGFP2 proteins. (C) Distribution patterns of the respective roGFP2-ORP1 proteins. Higher magnification view of po-roGFP2-ORP1 is shown. Note that high expression levels of the peroxisomal reporter proteins result in labelling of peroxisome tubules. (D) Box plot representations of the 400/480 nm fluorescence response ratios of the respective roGFP2 proteins. The bottom and top of each box represent the 25th and 75th percentile values, respectively; the horizontal line inside each box represents the median; and the horizontal lines below and above each box denote the mean minus and plus one standard deviation, respectively. The total number of measurements (two independent experiments; minimum 15 individual measurements in at least 20 randomly chosen cells) is indicated below each box plot. The data from the dMFF cell line were statistically compared with those from the CT cell line (**, p < 0.01). (E) Distribution patterns of pHRed-PO in control (C109) and MFF-deficient patient fibroblasts (MFFQ64⁎) at excitation wavelengths of 458 and 561 nm, along with digital visualisation of individual peroxisomal pH levels. Higher magnification views of boxed regions are indicated. (F) Calibration of the pHRed probe using cytosolic pHRed. The 458/561 ratiometric response is given at each pH level. AU, arbitrary units. (G) Quantification of peroxisomal pH in control (C109) and MFFQ64⁎ cells, converting the ratiometric response to pH using the calibration curve (n = 20). Scale bars, 10 μm; magnifications, 2 μm. Data are from at least 2–3 independent experiments. *, p < 0.05; ***, p < 0.001; two-tailed, unpaired t-test.