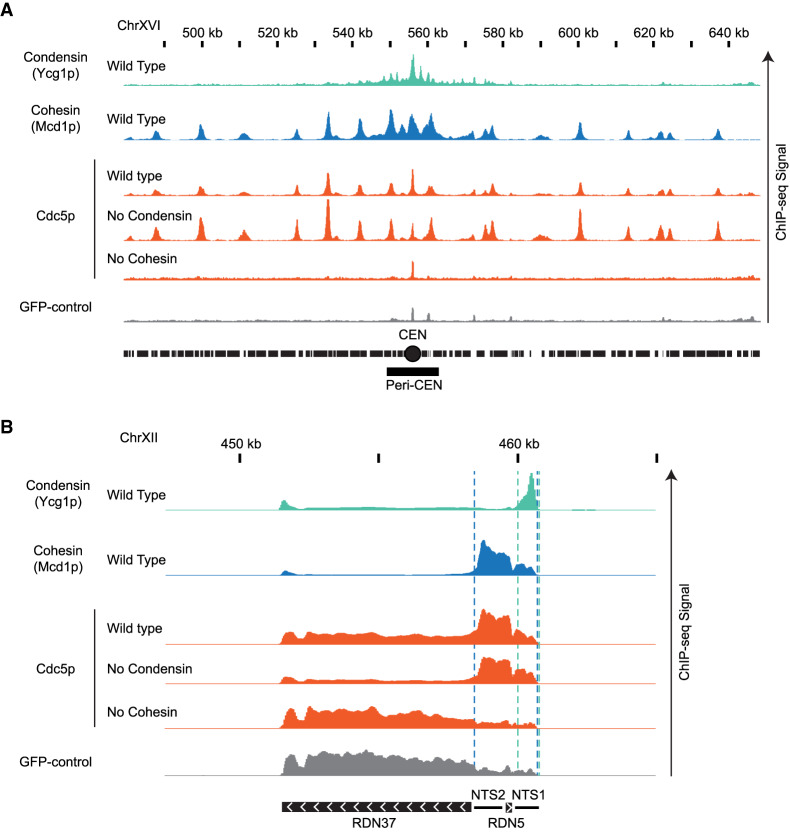
Figure 3.
Genome-wide localization of condensin, cohesin and Cdc5p. (A) Cohesin recruits Cdc5p at unique sequences genome-wide. ChIP-seq signals from a representative portion of chromosome XVI are presented with bar charts. The region presented includes the centromere (CEN; black circle) and the pericentric region (peri-CEN; black box). Signal coming from condensin subunit Ycg1p-Myc in wild-type strain is in green, signal from cohesin subunit Mcd1p in wild-type strain is in blue, signal coming from Cdc5p-Flag is in orange, and signal coming from GFP-control is in gray. Cdc5p ChIP-seq signal from wild type is in the first orange lane, from cells depleted of condensin subunit Brn1p-AID is in the second orange lane, and from cells depleted of cohesin subunit Mcd1p-AID is in the third orange lane. All strains were depleted of the indicated AID protein from G1 to mid-M and arrested in mid-M (staged mid-M). The scale is 0–15 for Ycg1p, 0–21 for Mcd1p, and 0–17 for Cdc5p. (B) Cohesin recruits Cdc5p at the repetitive RDN locus. ChIP-seq signals from a portion of chromosome XII are presented with bar charts. ChIP-seq signal from strains as in A. The region presented includes one copy of the rDNA repeat (9.1 kb) made of the transcribed regions RND37 and RND5 (black box with white arrows indicating the direction of transcription) and the nontranscribed regions NTS1 and NTS2 (black line). Region of enrichment of cohesin signal is marked by two blue dashed lines (from NTS2 to NTS1). Region of enrichment of condensin signal is marked by two green dashed lines (end of NTS1). The scale is 0–5200 for Ycg1p, 0–2300 for Mcd1p, and 0–1700 for Cdc5p.