Figure 1.
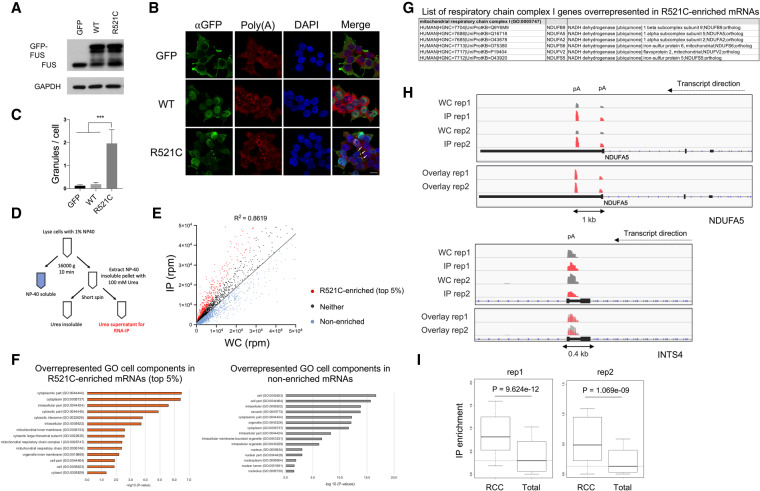
Nuclear-encoded respiratory chain complex (RCC) mRNAs are preferentially sequestered by ALS mutant FUS. (A) GFP-tagged FUS was transiently expressed in HEK293T cells for 48 h followed by Western blot of endogenous and GFP-tagged FUS. (B) Immunofluorescent staining of GFP-FUS and poly(A+) RNAs. Scale bar, 15 µm. (C) Quantification of GFP-FUS-poly(A+) RNA granules per cell. Total cells analyzed: N = 153. Fifty or more cells were analyzed in each transfection condition. (***) P < 0.001, Mann-Whitney U-test. (D) Flag-tagged mutant FUS-R521C was transiently expressed in U87 cells followed by fractionation to generate soluble and insoluble fractions. The insoluble fraction was extracted with 100 mM urea and the supernatant was IPed with an antibody targeting Flag-tagged FUS-R521C. Precipitated RNAs were purified and used for 3′READS to sequence poly(A+) RNAs bound by FUS-R521C. (E) Scatter plot of all poly(A+) RNAs associated with insoluble FUS-R521C (IP) versus poly(A+) RNA expression levels in whole cell (WC). Standard curve was plotted using linear regression. R521C-enriched poly(A+) RNAs are shown in red. RNAs with enrichment below the standard curve are nonenriched transcripts (pale blue). The remainders are neither (dark gray). (F) GO overrepresentation analysis of R521C-enriched and nonenriched transcripts in FUS-R521C IP. GO enrichment scores are displayed in −log10 (P-value). (G) List of respiratory chain complex I genes identified in overrepresentation analysis of R521C-enriched poly(A+) RNAs. (H) Genome browser snapshot of example gene read clusters from two replicates. (Top) NDUFA5. (Bottom) INTS4. (pA) Poly(A) site. (I) Box plots showing IP enrichment of RCC mRNAs (rep1: 64 genes, rep2: 60 genes) and total poly(A+) RNAs associated with FUS-R521C. Statistics: Wilcoxcon rank sum test.