Figure 7.
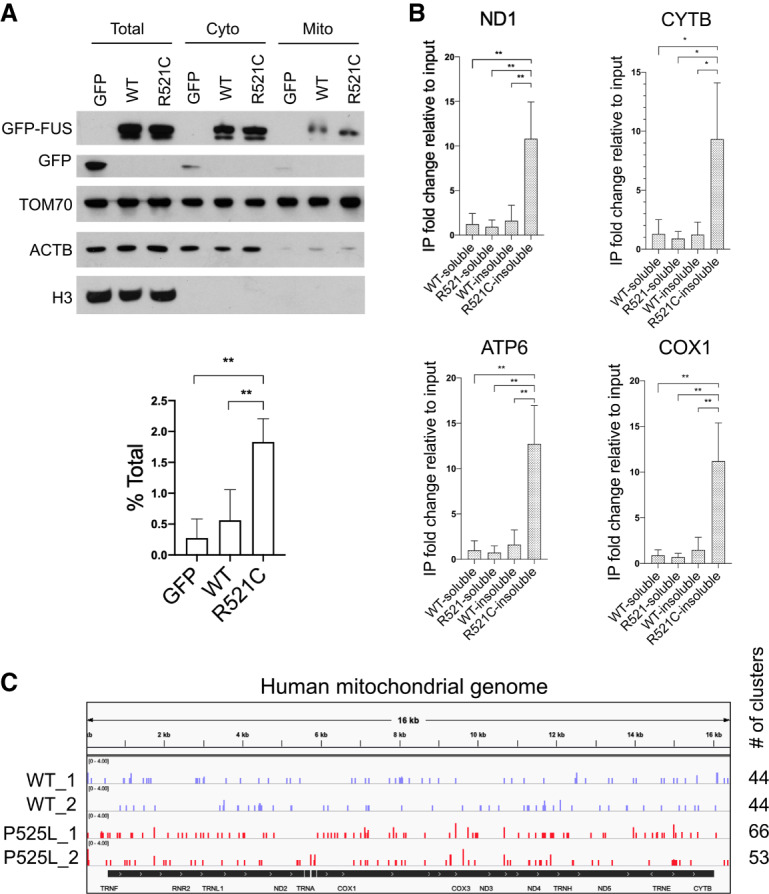
FUS associates with mitochondria and binds mitochondrial-encoded mRNAs. (A) HEK293T cells were transiently transfected with GFP-tagged wild-type and R521C mutant FUS for 48 h followed by mitochondrial fractionation. Western blot of mitochondrial fractionation. Purity of mitochondrial fractions was demonstrated by enrichment in mitochondrial protein TOM70 and absence of cytosolic protein beta actin and nuclear protein histone H3. Quantification of exogenously expressed GFP-tagged wild-type and FUS-R521C detected in mitochondrial fraction is displayed in percent of total protein. Data are shown in mean with SD from four independent experiments. (**) P < 0.01, unpaired t-test. (B) GFP-FUS IP followed by RT-qPCR analysis of RCC mRNAs encoded in mitochondrial genome from soluble fraction and urea-extracted insoluble fraction. Relative fold change is displayed in mean with SD from three independent experiments. (*) P < 0.05; (**) P < 0.01, unpaired t-test. (C) Realigned wild-type and P525L mutant FUS CLIP-seq data sets in iPSC-derived motor neurons (De Santis et al. 2019). Snapshot of wild-type and mutant FUS binding sites on mitochondrial-encoded transcripts. Number of read clusters aligned to mitochondrial genome was counted.
